1.0 Introduction
Over the past two decades, consumption of fresh vegetables has increased due to the fact that vegetables are been considered an important source of vitamins, nutrients and fibre [1,2]. In recent years, however, outbreaks of foodborne illness linked to vegetables have become more common [3,4], contaminated drinking water and irrigation with water contaminated by animal faeces discharged from sewage treatment facilities or surface runoff [5]. Vegetables are among the food groups implicated with greater frequency in recent years as causative agents of enteric diseases [6]. All types of fresh vegetables have the potential to harbour pathogens including but not limited to Shigella spp, Salmonella spp, enterotoxigenic and enterohaemorrhagic E. coli, Camphylobacter spp, Listeria monocytogenes, Clostridium botulinum are pathogens associated with foodborne illnesses [7].
Escherichia coli is one of the most frequent causes of diarrhoea in children in developing countries [8]. Diarrhoea illnesses are a severe public health problem and a major cause of morbidity and mortality in infants and young children [9]. Despite being harmless, various E. coli strains have acquired genetic determinants (virulence genes) giving them the capacity to cause illness for both humans and animals [10]. Infectious pathotypes of E. coli are related to the lack of sanitation, personal hygiene and also the consumption of vegetables harvested from contaminated rivers and irrigated farms[11] .
Escherichia coli pathotypes are collectively known as diarrheagenic E. coli (DEC). The DEC pathotypes differ regarding their preferential host colonization sites, virulence mechanisms and the ensuing clinical symptoms and consequences [12]. The DEC pathotypes can be classified as Entheropathogenic E. coli (EPEC), Enterohemorrhagic E. coli (EHEC/STEC), Enteroaggregative E. coli (EAEC), Enterotoxigenic E. coli (ETEC) and Enteroinvasive E. coli (EIEC). Each of these pathotypes represents a group of clones that share specific virulence factors [13].
In spite of increasing evidence that E. coli strains originate from human and animal faeces contain several virulence genes, only a few studies have investigated the presence of E. coli pathotype in irrigation farms [10,11,14,15]. The presence of E. coli strains with virulence genes profiles similar to EHEC, EPEC and ETEC in environmental water has already been reported [15], however, these environments that receive frequently domestic wastewater and mammalian faeces provide important resources for irrigation farms [15].
Unfortunately, these bacteria that are multidrug-resistant have been reported to have a high clinical and economic impact because of limitations in therapeutic options in the treatment of infections that they cause [7]. On these premises, the present study aimed at anti-microbial drug resistance profiling and molecular Detection of E. coli pathotypes isolated from irrigated spinach leaf (Amaranthus hybridus), waterleaf (Talinum triangulare) and pumpkin leaf (Telfairia occidentalis) in Keffi.
2.0 Materials and methods
2.1 Media, Reagents and Chemicals
Bacteriological media including MacConkey Agar (MCA), Mueller-Hilton Agar (MHA), Nutrient agar (NA), Luria Bertani (LB) broth, Eosine Methylene Blue (EMB) Agar, Nutrient Broth (NB), Simon Citrate Agar, Methyl red/Voges-Proskauer (MR/VP) medium and Peptone water (PW) were obtained from Oxoid Ltd, U.K. The crystal violet, Ethanol, Xylene solution, Creatinine, Pottasium hydroxide and Kovac’s reagents, were obtained from BDH chemical Ltd, England.
2.2 Antibiotic Discs
The antibiotic discs and potency including Amoxycillin (AMX) 10μg, Amoxicillin-clavulanic acid (AMT) 30μg, Cefotaxime(CTX) 30μg, Cefpodoxime (CPD) 10μg, Ceftazidime (CAZ) 30μg, Ceftriaxone(CRO) 30μg, Ciprofloxacin (CIP) 5μg, Co-cotrimoxazole (SXT) 25μg, Gentamicin (CN) 10μg, Ofloxacin (OFX) 5μg, Streptomycin (S) 10μgand Cefoxitin (FOX) 30 μg were obtained from Oxoid Ltd, U.K.
2.3 Description of the study area
The study area was Nasarawa State Keffi which is approximately 68km from Abuja, the Federal Capital Territory and 128km from Lafia, the Capital of Nasarawa State. Keffi is located between latitude 8o5 N of the equator and longitude 7o8 E and is situated at an altitude of 850 m above sea level.
2.4 Sample collection
A total of one hundred and thirty-five (135) samples of vegetables comprising of spinach leaf (Amaranthus hybridus), waterleaf (Talinum triangulare) and pumpkin leaf (Telfairia occidentalis) 45 samples of each vegetable were collected at irrigated farms in Keffi, namely Angwa Lambo, Market road and Old barracks, these farms are popular farms where irrigation is being practised in Keffi Nassarawa State, Nigeria. Samples were collected using alcohol sterile scissors. Labelled samples were transported to the Microbiology Laboratory, Nasarawa State University, Keffi in a sterile and sealed plastic bag for immediate analysis.
2.5 Isolation of Escherichia coli
E. coli were isolated from the vegetable samples following the method described by Bako, et al. [16]. The vegetables were cut into small sizes using a then transferred into enrichment broth (nutrient broth) and incubated for 18 hours then streak onto MacConkey Agar plates incubated at 37oC for 18 hours. The suspected E. coli growth with pinkish color on MacConkey plate was streaked onto Eosin methylene blue (EMB) agar plate and incubated at 37oC for 18hours for presumptive confirmation of E. coli. E. coli colonies with a metallic green sheen caused by large quantities of acid that is produced and that precipitate out the dyes onto the growth surface [16].
2.6 Screening for Diarrheagenic E. coli
Colonies were picked from the Eosin Methylene Blue (EMB) agar plates and homogenized in peptone water.1ml of the homogenized sample was diluted in 9ml Phosphate Buffered Saline (PBS) and spread plated on Sorbitol–MacConkey agar (SMAC) containing Cefixime(0.5 mg l−1) and incubated at 37°C for 24hrs. After24 hrs at 37°C, colonies that were pale/ colourless indicated the inability to ferment Sorbitol, which shows the presence of E. coli O157:H7 from foods that causes human diarrhoea.
2.7 Identification of Escherichia coli
The Escherichia coli isolates were further identify using standard microbiological procedures based on cultural, morphological and biochemical characteristics such as indole test, methyl red Test/Voges-Proskauer test and citrate test as described in previous study [17,18]
2.8 Antibiotic susceptibility test:
The antibiotic susceptibility test of the bacterial isolates was carried out as earlier described by the Clinical and Laboratory Standards Institute [19]. Briefly, three (3) pure colonies of isolated bacteria species from vegetables were inoculated into 5ml sterile 0.85% (w/v) NaCl (normal saline) and the turbidity of the bacteria suspension was adjusted to the turbidity equivalent to 0.5 McFarland’s standard. The McFarland’s standard was prepared as follows; 0.5ml of 1.172% (w/v) BaCl2.2H20 was added into 99.5ml of 1% (w/v) H2SO4. A sterile swab stick was soaked in standardized bacteria suspension and streaked on Mueller Hilton agar plates and the antibiotic discs were aseptically placed at the centre of the plates and allowed to stand for 1 h for a pre-diffusion time. The plates were incubated at 370C for 24h. The diameter zone of inhibition was measured and the result of the susceptibility was interpreted in accordance with the susceptibility breakpoint earlier described by the Clinical and Laboratory Standards Institute [19].
2.9 Molecular Detection of E. coli Pathotypes
2.9.1 Amplification of Escherichia coliGenes
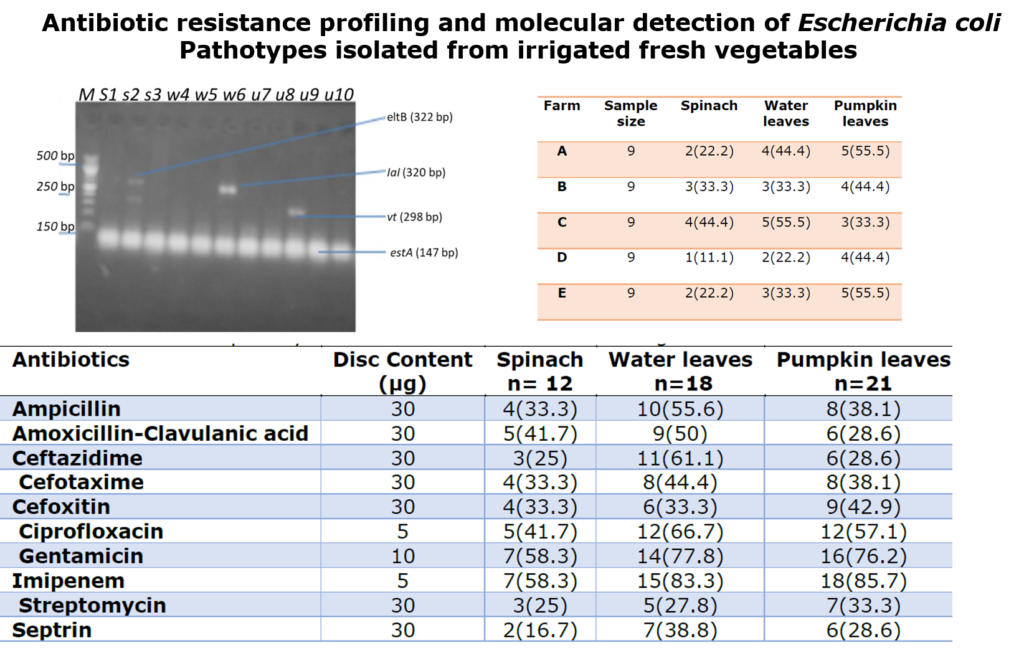
The amplification of DEC genes was done by PCR assay of the DNA extracted from E. coli isolates as described by Nguyen et al. [20]. The DNA templates were subjected to multiplex PCR with specific primers for the detection of the following virulence markers: eaeA (structural gene for intimin of EHEC and EPEC), bfpA (structural gene for the bundle-forming pilus of EPEC), vt1 and/or vt2 (Shiga toxins 1 and 2 of EHEC), eltB and/or estA (enterotoxins of ETEC), ial (invasion-associated locus of the invasion plasmid found in EIEC and Shigella) and pCVD (the nucleotide sequence of the EcoRI-PstI DNA fragment of pCVD432 of EAEC). Primers and amplicon size of E. coli pathotypes described in previous study [21] was used (Table 1)
The PCR was performed with a 25 μl reaction mixture containing 5 μl of template DNA, 0.2 μl of 18x PCR buffer II, 1.6 μl of a 1.25 mM mixture of deoxynucleoside triphosphates, 1.6 μl of 25 mM MgCl2, 0.1 μl of 5 U of AmpliTaq Gold DNA polymerase per μl and a 0.2 μM concentration of each primer except primer VT1, which was used at a concentration of 0.4 μM. The thethermocycling conditions used are as follows: 95°C for 5 min (Initial denaturation), 94°C for 20 sec. (denaturation) 55°C for 30 sec. (Annealing) and 72°C for 30 sec. (initial extension) for 30 cycles, with a final 7 min extension at 72°C [20].
2.9.2 Agarose Gel Electrophoresis
The agarose gel electrophoretic assay for detection of amplified genes for different DEC pathotypes was carried out as described by Nguyen et al. [20]. Briefly, 8μl of PCR products stained with ethidium bromide was loaded into 1.0% (wt/vol) agarose gel wells with a molecular marker run concurrently at 120 V for 30 min. The DNA bands were visualized and photographed under UV light 595nm.
Table 1: Primers and amplicon size of Escherichia coli pathotypes that was used
| Primer | Target gene | Primer sequence | Amplicon size (bp) |
| LT | eltB | 5̍-TCTCTATGTGCATACGGAGC-3̍ 5̍-CCATACTGATTGCCGCAAT-3̍ | 322 |
| ST | estA | 5̍-GCTAAACCAGTAGAGGTCTTCAAAA-3̍ 5̍-CCCGGTACAGAGCAGGATTACAACA-3̍ | 147 |
| VT1 | vt1 | 5̍-GAAGAGTCCGTGGGATTACG-3̍ 5̍-AGCGATGCAGCTATTAATAA-3̍ | 130 |
| VT2 | vt2 | 5̍-ACCGTTTTTCAGATTTTGACACATA-3̍ 5̍-TACACAGGAGCAGTTTCAGACAGT-3̍ | 298 |
| Eae | eaeA | 5̍-CACACGAATAAACTGACTAAAATG-3̍ 5̍-AAAAACGCTGACCCGCACCTAAAT-3̍ | 376 |
| SHIG | Ial | 5̍-CTGGTAGGTATGGTGAGG-3̍ 5̍-CCAGGCCAACAATTATTTCC-3̍ | 320 |
LT= Enterotoxigenic E. coli (ETEC); ST=Enterotoxigenic E. coli (ETEC);VT=Enterohemorrhagic E.coli (EHEC); Eae=Enterohemorrhagic E.coli (EHEC); SHIG=Enteroinvasive E.coli (EIEC); BfpA=Enteropathogenic E.coli (EPEC); EA= Enteroaggregative E.coli (EAEC)
3.0 Results
3.1 Cultural, morphological and biochemical characteristics of Escherichia coli isolates
The cultural, morphological and biochemical characteristics of Escherichia coli isolated from vegetables leaves from Farms in Keffi revealed a pinkish colony on MCA which grew with greenish metallic sheen on EMB agar, gram negative rod and had biochemical reactions namely: indole-positive, methyl red-positive, Voges-Proskauer-negative, citrate-negative and Ornithine –positive all indicated E. coli.
3.2 Percentage occurrence of the Escherichia coli isolates
The percentage occurrence of the Escherichia coli isolates is as given in Table 2. out of 135 samples collected the percentage occurrence was 37.7% and the highest percentage occurrence was observed from pumpkin leaves with 46.6% followed by waterleaves which had 40.0% and the lowest was spinach with 26.6% respectively.
The occurrence of Escherichia coli isolated from vegetable leaves in respect to different farms in Keffi is as given in Table 3. It was observed that from farm A spinach had 22.2%, water leaves had 44.4% and pumpkin leaves had 55.5%. Farm B spinach had 33.3%, waterleaves had 33.3% and pumpkin leaves had 44.4% respectively. Farm C spinach had 44.4%, waterleaves had 55.5% and pumpkin leaves had 33.3%. Farm D spinach had 11.1%, waterleaves had 22.2% and pumpkin leaves had 44.4% and from E spinach had 22.2%, waterleaves had 33.3% and pumpkin leaves had 55.5% respectively.
Table 2: Percentage occurrence of Escherichia coli isolated from vegetable leaves from farms in Keffi
| Vegetables | No. of Samples | No. (%) isolated |
| Spinach | 45 | 12(26.6) |
| Water leaves | 45 | 18(40.0) |
| Pumpkin leaves | 45 | 21(46.6) |
| Total | 135 | 51(37.7) |
Table 3: Occurrence of Escherichia coli isolated from vegetable leaves in respect to different farms in Keffi
| Farm | Sample size | Spinach | Water leaves | Pumpkin leaves |
| A | 9 | 2(22.2) | 4(44.4) | 5(55.5) |
| B | 9 | 3(33.3) | 3(33.3) | 4(44.4) |
| C | 9 | 4(44.4) | 5(55.5) | 3(33.3) |
| D | 9 | 1(11.1) | 2(22.2) | 4(44.4) |
| E | 9 | 2(22.2) | 3(33.3) | 5(55.5) |
3.3 Antibiotic susceptibility of Escherichia coli isolated from vegetable leaves
The antibiotic susceptibility of Escherichia coli isolated from vegetable leaves in respect to different vegetables is as given in Table 4 Shows that Escherichia coli isolated from spinach more susceptible to gentamicin and imipenem with 58.3% followed by ciprofloxacin and amoxicillin-clavulanic acid with 41.7%, ampicillin and cefoxitin had 33.3%, ceftazidime and streptomycin had 25.0% and the lowest was septrin with 16.7%. Escherichia coli isolated from water leaves were highly susceptible to imipenem with 83.3% followed by gentamicin with 77.8%, ciprofloxacin with 66.7%, ceftazidime with 61.1%, ampicillin with 55.6%, amoxicillin-clavulanic acid with 50.0%, cefotaxime with 44.4%, cefoxitin with 33.3%, septrin with 38.8% and the lowest was streptomycin which had 25.0%.
It was observed that Escherichia coli isolated from pumpkin leaves were highly susceptible to imipenem with 85.7% followed by gentamicin with 76.2%, ciprofloxacin with 57.2% but less susceptible to Cefoxitin with 42.9%, ampicillin and Cefotaxime had 38.1%, Streptomycin with 33.3%, Amoxicillin-Clavulanic acid, Ceftazidime and Septrin had the lowest with 28.6% respectively.
Table 4: Antibiotics susceptibility of Escherichia coli isolated from vegetable leaves from farms in Keffi
| Antibiotics | Disc Content (µg) | Spinach n= 12 | Water leaves n=18 | Pumpkin leaves n=21 |
| Ampicillin | 30 | 4(33.3) | 10(55.6) | 8(38.1) |
| Amoxicillin-Clavulanic acid | 30 | 5(41.7) | 9(50) | 6(28.6) |
| Ceftazidime | 30 | 3(25) | 11(61.1) | 6(28.6) |
| Cefotaxime | 30 | 4(33.3) | 8(44.4) | 8(38.1) |
| Cefoxitin | 30 | 4(33.3) | 6(33.3) | 9(42.9) |
| Ciprofloxacin | 5 | 5(41.7) | 12(66.7) | 12(57.1) |
| Gentamicin | 10 | 7(58.3) | 14(77.8) | 16(76.2) |
| Imipenem | 5 | 7(58.3) | 15(83.3) | 18(85.7) |
| Streptomycin | 30 | 3(25) | 5(27.8) | 7(33.3) |
| Septrin | 30 | 2(16.7) | 7(38.8) | 6(28.6) |
3.4 Occurrence of pathotypes genes in antibiotic-resistant E. coli from vegetable leaves
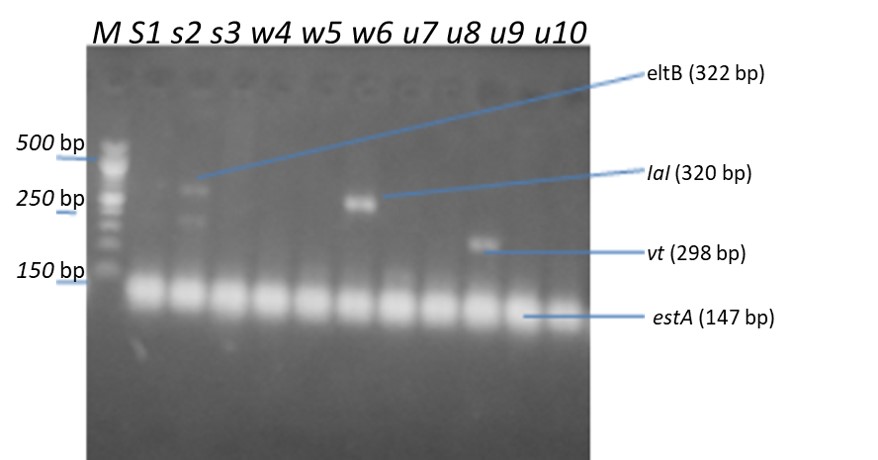
The pathotypes genes detected in antibiotic-resistant Escherichia coli from vegetable leaves are as given in Table 5 and Figure 1. Results revealed the presence of eltB pathotype base pair of 322bp, IaI pathotpye base pair of 320bp, vt2 pathotype base pair of 298bp and estA pathotype of 147bp. The E. coli pathotype that was detected were eltB (33%) from spinach only, IaI pathotype from spinach and water leaves each (33%), vt2 from pumpkin leaves (33%) and estA from spinach, water leaves and pumpkin leaves each (100%).
Table 5: Occurrence of pathotypes genes in Antibiotic-resistant E. coli from the vegetable leaves
| Pathotypes genes | Spinach (n=3) | Waterleaves (n=3) | Pumpkin leaves (n=4) |
| eltB | 1(33.3) | 0(00.0) | 0(00.0) |
| Ial | 1(33.3) | 1(33.3) | 0(00.0) |
| vt2 | 0(00.0) | 0(00.0) | 1(33.3) |
| estA | 3(100) | 3(100) | 4(100) |
4.0 Discussion
The consumption of fresh vegetables has increased notably in recent years due to multiple contributions of nutrients and functional properties [22]. A diet rich in vegetables has been shown to protect against various types of cancer and chronic illnesses, such as coronary heart disease [23-25]. However, at the same time, the consumption of fresh produce is associated with a growing number of foodborne outbreaks due to bacterial contamination of these products. Leafy greens, such as lettuce, spinach and fresh herbs are some of the vegetables most frequently linked to bacterial infections [26].
Foodborne illness may be the cause of fresh produce contamination by pathogenic bacteria. This contamination may originate from manure, soil, sewage, surface water, or wildlife. It may also occur during washing, slicing, soaking, packing, and food preparation. Among the bacteria associated with foodborne illnesses are Listeria monocytogenes and E. coli [26].
The percentage occurrence of E. coli from the different vegetable farms in Keffi was high with37.7% but lower than the study reported by Karaye et al. [27] in Zaria, Bako et al. [16] in Kaduna with a prevalence rate of 47.6%. The occurrences of E. coli from the different vegetable samples were in agreement with the studies that have been reported by different authors; Kabiru et al. [28] in Zaria, Reuben and Makut, [29] in Lafia and Karayeet al. [27] in Zaria.
The high occurrence of E.coli as observed in this research could be attributed to high microbial load on vegetables especially those grown around faecal waste dumpsite during the dry season when there is no rainstorm washing to reduce the microbial load on the vegetables as reported by Karaye et al. [27], and Kabiru et al. [28] also opined that manure application and irrigation with contaminated water as frequently done during the dry season in Keffi could have been another reason for this finding in which isolates were confirmed highly E.coli positive.
It was observed in this study that E.coli was isolated more from ugu leaves followed by waterleaves from different farms studied or sampled and this is similar to the findings of Reuben and Makut [29]. The isolation of the pathogen E. coli from vegetable farms in Keffi suggests that contamination of spinach, water leaves and ugu exposes populates to food-borne hazards. The socio-economic implication is that resources and time are wasted on medication. The effects of food-borne infections are also greatly felt by immune-compromised individuals such as, pregnant women, children, diabetic patients Bako et al. [16].
The antibiotic susceptibility of Escherichia coli isolated from vegetable leaves from farms in Keffi showed that the E. coli from different farms were more susceptible to gentamicin and imipenem and ciprofloxacin but less susceptible to amoxicillin-clavulanic acid, ampicillin, cefoxitin, ceftazidime, streptomycin and septrin. This is not in agreement with studies reported by Obinna and Destiny, [30]. This observation presents a direct challenge to public health, where this E. coli could serve as a gene pool for horizontal gene spread of antibiotic resistance genes to others when coming in contact with them.
5.0 Conclusion
The findings of this research have indicated the occurrence of Escherichia coli isolated from different vegetable samples namely spinach, waterleaves and pumpkin leaves collected from vegetable farms in Keffi. The percentage occurrence was high in pumpkin leaves samples than in the other two vegetables sampled. The Escherichia coli isolated were more susceptible to imipenem, ciprofloxacin and gentamicin but less susceptible to Streptomycin, Amoxicillin-Clavulanic acid, Ceftazidime and Septrin. Different E. coli Pathotype genes were detected. This study presents a direct challenge to public health, where E. coli could serve as a gene pool for horizontal gene spread of antibiotic resistance genes
Funding: This research did not receive any funding grant
Author’s contributions: All authors contributed in preparing this article.
Conflict of interest: The authors declared no conflict of interest.
Acknowledgements: Not Applicable
References
1. Ramos, B.; Miller, F.; Brandão, T.; Teixeira, P.; Silva, C. Fresh fruits and vegetables—an overview on applied methodologies to improve its quality and safety. Innovative Food Science & Emerging Technologies 2013, 20, 1-15.
2. Tsado, A.; Lawal, B.; Santali, E.; Shaba, A.; Chirama, D.; Balarabe, M.; Jiya, A.; Alkali, H. Effect of different processing methods on nutritional composition of Bitter Leaf (Vernonia amygdalina). IOSR Journal of Pharmacy 2015, 5.
3. Paggi, M.S. An assessment of food safety policies and programs for fruits and vegetables: Food-borne illness prevention and food security; 2008.
4. Warriner, K.; Huber, A.; Namvar, A.; Fan, W.; Dunfield, K. Recent advances in the microbial safety of fresh fruits and vegetables. Advances in food and nutrition research 2009, 57, 155-208.
5. Wachtel, M.R.; Whitehand, L.C.; Mandrell, R.E. Association of Escherichia coli O157: H7 with preharvest leaf lettuce upon exposure to contaminated irrigation water. Journal of food protection 2002, 65, 18-25.
6. Berger, C.N.; Sodha, S.V.; Shaw, R.K.; Griffin, P.M.; Pink, D.; Hand, P.; Frankel, G. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environmental microbiology 2010, 12, 2385-2397.
7. Akinde, S.B.; Sunday, A.A.; Adeyemi, F.M.; Fakayode, I.B.; Oluwajide, O.O.; Adebunmi, A.A.; Oloke, J.K.; Adebooye, C.O. Microbes in irrigation water and fresh vegetables: potential pathogenic bacteria assessment and implications for food safety. Applied Biosafety 2016, 21, 89-97.
8. Black, R.E.; Brown, K.H.; Becker, S.; Alim, A.A.; Merson, M.H. Contamination of weaning foods and transmission of enterotoxigenic Escherichia coli diarrhoea in children in rural Bangladesh. Transactions of the Royal Society of Tropical Medicine and Hygiene 1982, 76, 259-264.
9. Vilchez, S.; Reyes, D.; Paniagua, M.; Bucardo, F.; Möllby, R.; Weintraub, A. Prevalence of diarrhoeagenic Escherichia coli in children from Leon, Nicaragua. Journal of medical microbiology 2009, 58, 630-637.
10. Kambire, O.; Adingra, A.A.; Yao, K.M.; Koffi-Nevry, R. Prevalence of virulence genes associated with diarrheagenic pathotypes of Escherichia coli isolates from water, sediment, fish, and crab in Aby Lagoon, Côte d’Ivoire. International journal of microbiology 2017, 2017.
11. Aijuka, M.; Santiago, A.E.; Girón, J.A.; Nataro, J.P.; Buys, E.M. Enteroaggregative Escherichia coli is the predominant diarrheagenic E. coli pathotype among irrigation water and food sources in South Africa. International journal of food microbiology 2018, 278, 44-51.
12. Gomes, T.A.; Elias, W.P.; Scaletsky, I.C.; Guth, B.E.; Rodrigues, J.F.; Piazza, R.M.; Ferreira, L.; Martinez, M.B. Diarrheagenic escherichia coli. brazilian journal of microbiology 2016, 47, 3-30.
13. Nataro, J.P.; Kaper, J.B. Diarrheagenic escherichia coli. Clinical microbiology reviews 1998, 11, 142-201.
14. Castro-Rosas, J.; Cerna-Cortés, J.F.; Méndez-Reyes, E.; Lopez-Hernandez, D.; Gómez-Aldapa, C.A.; Estrada-Garcia, T. Presence of faecal coliforms, Escherichia coli and diarrheagenic E. coli pathotypes in ready-to-eat salads, from an area where crops are irrigated with untreated sewage water. International journal of food microbiology 2012, 156, 176-180.
15. Shah, M.S.; Eppinger, M.; Ahmed, S.; Shah, A.A.; Hameed, A.; Hasan, F. Multidrug-resistant diarrheagenic E. coli pathotypes are associated with ready-to-eat salad and vegetables in Pakistan. Journal of the Korean Society for Applied Biological Chemistry 2015, 58, 267-273.
16. Benjamin, B.; Uba, A.; Yusha’u, M.; Maikaje, D.; Nyakaat, N.N.; Daniel, A.M. Isolation of Escheria coli from fruits and vegetables in Kaduna Metropolis. International Journal of Engineering Science 2018, 18598.
17. Makut, M.; Madaiki, K.; Obiekezie, O. Molecular characterization of xanthan gum producing Xanthomonas Campestris isolated from dark rot spotted leaves in Keffi, Nasarawa State, Nigeria. AROC in Pharmaceutical and Biotechnology 2022, 2, 01-08, doi:10.53858/arocpb02010108.
18. Olunrebi, B.; Onaolapo, J.; Bolaji, R.O.; Otaru, S. Antimicrobial Susceptibility Pattern of Enteric Bacteria from Fresh cow milk and handlers in Zaria Metropolis, Kaduna State Nigeria. AROC in Pharmaceutical and Biotechnology 2021, 01, 35-42, doi:10.53858/arocpb01023542.
19. Kassim, A.; Omuse, G.; Premji, Z.; Revathi, G. Comparison of Clinical Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing guidelines for the interpretation of antibiotic susceptibility at a University teaching hospital in Nairobi, Kenya: a cross-sectional study. Annals of clinical microbiology and antimicrobials 2016, 15, 1-7.
20. Nguyen, T.V.; Le Van, P.; Le Huy, C.; Gia, K.N.; Weintraub, A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. Journal of clinical microbiology 2005, 43, 755-760.
21. Toma, C.; Lu, Y.; Higa, N.; Nakasone, N.; Chinen, I.; Baschkier, A.; Rivas, M.; Iwanaga, M. Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. Journal of clinical microbiology 2003, 41, 2669-2671.
22. Park, S.; Park, S.-Y. Can antioxidants be effective therapeutics for type 2 diabetes? Yeungnam University Journal of Medicine 2021, 38, 83.
23. Lawal, B.; Shittu, O.K.; Oibiokpa, F.I.; Berinyuy, E.B.; Mohammed, H. African natural products with potential antioxidants and hepatoprotectives properties: a review. Clinical Phytoscience 2016, 2, 23, doi:10.1186/s40816-016-0037-0.
24. Onikanni, A.S.; Lawal, B.; Olusola, A.O.; Olugbodi, J.O.; Sani, S.; Ajiboye, B.O.; Ilesanmi, O.B.; Alqarni, M.; Mostafa-Hedeab, G.; Obaidullah, A.J., et al. Sterculia tragacantha Lindl Leaf Extract Ameliorates STZ-Induced Diabetes, Oxidative Stress, Inflammation and Neuronal Impairment. J Inflamm Res 2021, 14, 6749-6764, doi:10.2147/jir.s319673.
25. Onikanni, A.S.; Lawal, B.; Oyinloye, B.E.; Mostafa-Hedeab, G.; Alorabi, M.; Cavalu, S.; Olusola, A.O.; Wang, C.-H.; Batiha, G.E.-S. Therapeutic efficacy of Clompanus pubescens leaves fractions via downregulation of neuronal cholinesterases/Na+-K+ATPase/IL-1 β, and improving the neurocognitive and antioxidants status of streptozotocin-induced diabetic rats. Biomedicine & Pharmacotherapy 2022, 148, 112730, doi:https://doi.org/10.1016/j.biopha.2022.112730.
26. Luna-Guevara, J.J.; Arenas-Hernandez, M.M.; Martínez de la Peña, C.; Silva, J.L.; Luna-Guevara, M.L. The role of pathogenic E. coli in fresh vegetables: Behavior, contamination factors, and preventive measures. International journal of microbiology 2019, 2019.
27. Karaye, G.; Karaye, K.; Kaze, P. Detection of Escherichia Coli in Freshly Harvested Spinach Samples Collected from Five Different Markets in Zaria. J Microbes Microbio Techni 2019, 2, 101.
28. Kabiru, L.M.; Bello, M.; Kabir, J.; Grande, L.; Morabito, S. Detection of pathogenic Escherichia coli in samples collected at an abattoir in Zaria, Nigeria and at different points in the surrounding environment. International journal of environmental research and public health 2015, 12, 679-691.
29. Reuben, C.; Makut, M. Occurrence of Escherichia coli O157: H7 in vegetables grown and sold in Lafia metropolis, Nigeria. World Journal of Microbiology 2014, 1, 017-021.
30. Obinna, C.N.; Destiny, N. Research article antibiogram of bacteria species isolated from vegetables in Ado-Odo Ota, Nigeria. J. Biol. Sci 2016, 16, 188-196.