1.0 Introduction
Klebsiella (K.) pneumoniae is a Gram-negative opportunistic nosocomial bacterial pathogen. It is involved in several localized and disseminated hospital-acquired infections such as burns infections, sepsis, respiratory and gastrointestinal tract infections, urinary tract infections, pyogenic liver abscesses, and soft tissue and wound infection [1]. The emergence of carbapenem-resistant K. pneumoniae (CRKP) strains has become an ultimate challenge for public health globally due to their ability to disseminate rapidly in the hospital environment and their extended antibiotic resistance phenotypes [2].
In 2001, the first K. pneumoniae isolate with KPC-2 production was identified in the USA. There are many mechanisms in K. pneumoniae that can drive carbapenem resistance; KPC-2 production is just one [3]. A few years later, outbreaks began to appear in several countries. Nowadays, it is the most common carbapenemase-producing Enterobacterales (CPE), and is considered one of the most rapidly growing global threats due to the high mortality in hospital-associated infections [4].
In 2017, CRKP was classified among those global critical pathogens listed by WHO concerning discovering and developing new antibiotics [5]. In Nigeria, K. pneumoniae was among the most common causes of lower respiratory tract infections, neonatal septicemia, and bacteremia in children [6, 7]. Infections with multidrug-resistant (MDR) pathogens impose a significant and increasing burden on both patients and healthcare providers [8]. Among MDR pathogens, Klebsiella pneumoniae (K. pneumonia or KP) is one of the world’s most dangerous superbugs; and becoming resistant to virtually every antibiotic available today [9].
Carbapenem class of antibiotics consists of highly effective agents commonly used for the treatment of severe or high-risk bacterial infections [10]. This class of antibiotics is usually reserved for known or suspected MDR bacterial infections[11]. Similar to penicillins and cephalosporins, carbapenems are members of the βeta-lactam class of antibiotics, which kill bacteria by binding to penicillin binding proteins, thus inhibiting bacterial cell wall synthesis [12].
Many countries have experienced a dramatic upswing in the prevalence of Enterobacteriaceae that produce both extended spectrum beta-lactamases (ESBLs) and carbapenemases such as the Klebsiella pneumoniae carbapenemases (KPC) [13, 14]. As of 2013, 70% of Klebsiella pneumoniae isolates are resistant to third generation cephalosporins and 60% are resistant to carbapenems [15]. The growing resistance and difficulty of treating such multi-drug resistance Enterobacteriaceae has led to the renaissance of the use of antibiotics such as colistin, which was discovered in the 1950s but rarely used until recently due to the unattractive levels of toxicity [16, 17]
Unfortunately, no information or data about the incidence or otherwise is available in Nigeria despite the fact that it is a significant health problem even in the developed world. Therefore, the aim of the present study is to carryout molecular detection of carbapenemnase resistance in Klebsiella pneumoniae isolated from urine of patients assessing General Hospital in Keffi, Nasarawa state, Nigeria
2.0 Materials and Methods
2.1 Media and Reagents
Nutrient Agar (NA), Nutrient Broth (NB), Xylose Lysine Deoxycholate (XLD) agar, MacConkey Agar (MCA), Mueller-Hilton Agar (MHA), Simmons Citrate Agar (SCA) Methyl Red (MR), Voges-Proskauer (VP), Peptone Water (PW). BashingBead™ Buffer (Zymo Research, Made in USA), Genomic Lysis Buffer (Zymo Research, Made in USA), DNA Pre-Wash Buffer (Zymo Research, Made in USA), g-DNA Wash Buffer (Zymo Research, Made in USA), DNA Elution Buffer (Zymo Research, Made in USA), Sterile Nuclease – free water (VWR, Solon, OH), OneTaq® Quick-Load® 2X. Master Mix (New England BioLabs,), Quick-Load® Purple 100 bp DNA Ladder (New England BioLabs,), Crystal Voilet, Kovac’s reagent, Potassium hydroxide, beta-naphthol.
2.2 Antibiotics
Amoxicillin (AML:10 µg), Ampicillin (AMP:10 µg), Cefexime (CFM:5 µg), Ceftriaxone (CRO:30 µg), Ciprofloxacin (CIP:5 µg), Gentamicin (CN:30 µg), Imipenem (IMP:10 µg), Streptomycin (S:30 µg) were from Oxoid, Ltd (U.K).
2.3 Study Area and Location
The study area is Keffi metropolis is located between longitude 8-5 So and latitude 7oN and above the sea level of 630m. Keffi is approximately 53km away from the Federal Capital Territory, Abuja and 128km away from the state capital Lafia [18]. This study was carried out in Keffi, Nasarawa State, Nigeria General Hospital, Keffi.
2.4 Sample Collection
The ethical approval was obtained from the Ministry of Health Lafia. A total number of 210 urine samples was collected from General Hospital Keffi and then transported using ice box to the Microbiology laboratory in Nasarawa State University, Keffi, for analysis.
2.5 Isolation of Klebsiella pneumoniae
The K. pneumoniae was isolated from the urine samples by modification of the method earlier described Cheesbrough, [19]. A loopful was streaked across MacConkey agar and incubated at 37°C for 24 h. Pink mucoid colonies on MacConkey was selected and cultured on Xylose Lysine Deoxycholate (XLD) agar. Yellow colonies formed on XLD agar which was taken as suspect K. pneumoniae.
2.6 Identification of Klebsiella pneumoniae
The Identification of Klebsiella pneumoniae was done using the commercial biochemical kit (KB003 H125™). Gram staining of suspect organisms was carried out as described previously by Cheesbrough, [19]. The presumptive K. pneumoniae isolates that were Gram negative, rod shape, indole negative, methyl red negative, citrate positive, and Voges-Proskauer negative were confirmed using KB003 H125 Kit following the manufacturer’s instruction
2.7 Antibiotics Susceptibility Testing
The antimicrobial susceptibility testing of the K. pneumoniae isolates were carried out as earlier described by Clinical and Laboratory Standard Institute [20]. Pure colonies of K. pneumoniae isolates were inoculated into 5 ml sterile 0.85% (w/v) NaCl (normal saline) and the turbidity of the isolates was adjusted to the turbidity equivalent to 0.5 McFarland’s standard. The McFarland’s standard was prepared as follows: 0.5ml of 1.172% (w/v) BaCl2H2O was added into 99.5ml of 1% (w/v) H2S04. A sterile swab stick was soaked in the standardized bacteria suspension and streaked on Mueller-hinton agar figures so as to have influent growth and antibiotics disc were asceptically placed at the centre of the figures and allowed to stand for 1 hour for pre-diffusion was used to evaluate the susceptibility or resistance of K. pneumoniae isolates against Ciprofloxacin 5 µg , Streptomycin 10 µg , Trimethoprim 25 µg, Gentamicin 30 µg ,Imipenem 10 µg , Ceftriaxone 30 µg, Cefixime 5 µg, Amoxicillin 10µg, Ampicillin 10µg (Oxoid, UK). The figures were incubated at 37°C for 24 h. After incubation, the inhibition zones were measured and interpreted by the recommendations of the Clinical and Laboratory Standards Institute.
2.8 Determination of Multiple Antibiotics Resistance (MAR) Index of the isolates
The MAR index of the antibiotics resistant isolates was determined using the formula MAR Index = No. antibiotics isolates was resistant to / No. of antibiotics tested as described previously [21]
2.9 Classification of Antibiotics Resistance of the isolates
Antibiotic resistance in the isolates was classified into: multi drug resistance (MDR), extensive drug resistance (XDR), pan drug resistance (PDR) and non-multi drug resistance (NMDR) [22]
2.10 DNA Extraction
Purification of K. pneumonia was done on MacConkey agar, and following Zymoresearch, 2018 manufacturer’s instruction K. pneumoniae was isolated from a 24hours culture. Estimation of concentration, purity and yield of DNA sample was carried out using absorbance method with the spectrophotometer (Nanodrop 1000). DNA purity was estimated by calculating the A260/A280 ratio [23] and this was done with spectrophotometer’s computer software (where A260/A280 ratio ranges from 1.7-1.9)
2.11 DNA Amplification
The presence of genes (KPC, VIM, SPM, IPM and OXA) were tested using primer sets and conditions listed in Table below. PCR cocktail of 25μl was prepared (4μl of DNA, 2μl of forward primer, 2μl of reverse primer, 12.5μl of Master mix, 4.5μl of Nuclease free sterile H2O)in tubes then placed in a thermocycler (Thermofisher Scientific, Finland) Initial denaturation temperature 95℃ for 5min, 35 cycles of amplification took place at second denaturation at 94℃ for 45min, annealed at 60℃ for 40sec, extended at 35℃ 1min, final extension at 72℃ for 5min and hold 4℃. The primers sequence and amplicon size for carbapenemase resistant genes are presented in table below.
blaSPM (271)= F: AAAATCTGGGTACGCAAACG
R: ACATTATCCGCTGGAACA
blaVIM (502)= F: GTGTTTGGTCGCATATCGCAA
R: ATTCAGCCAGATCGGCATCGG
blakpc (924)= F: GGTTTGGCGATCTGGTTTTC
R: CGGAATGGCTCATCACGATC
2.12 Agarose Gel Electrophoresis
Ten microliter of the PCR products and 10 μl DNA ladder (Purple 100 bp DNA Ladder, New England BioLabs) was analyzed by electrophoresis on 1% TAE agarose gel containing 0.5 μl of ethidium bromide pipetted into well created with comb. Electrophoresis was run at 100 volts for 1 hour, after which DNA amplicons were then viewed on a UV trans-illuminator.
3.0 Results
3.1 Isolation and Identification of Klebsiella pneumoniae
The cultural, morphological, and biochemical characteristics Klebsiella pneumoniae from isolated from urine of patients is as given in Table 1 Pinkish with mucoid colony on MCA which grew with yellowish on XLD agar, was Gram negative rod and biochemical reactions namely: Indole negative, methyl red negative, Voges-Proskauer positive, Citrate positive, ONPG- negative indicated K. pneumoniae
Table 1: Cultural, Morphological and Biochemical characteristics of Klebsiella pneumoniae isolates.
| Cultural characteristics | Morphological characteristics | Biochemical Characteristics | |||||||||||
| Gram reaction | Morphology | IND | MR | VP | CT | ONPG | ORN | UR | H2S | ||||
| Pink and mucor colony on MCA and yellowish on XLD agar | – | Rod | – | – | + | + | – | – | – | – |
+ = Positive, – = negative, IND = Indole; MR = Methyl red; VP= Voges-Proskauer, CT = Citrate, ORN = Ornithine; ONPG = Ortho-Nitrophenyl-β-galactosidase, UR = Urease, H2S = Hydrogen Sulphide, Mal = Malonate
3.2 Occurrence of Klebsiella pneumoniae
The occurrence of Klebsiella pneumoniae isolates were recorded 49(23.3%) out of the 210 urine samples. The occurrences of K. pneumoniae in relation to age and gender of the patients are shown in Table 2. The highest occurrence of K. pneumoniae in the patients were observed at age 21-30 (27.1%) followed by 31-40 (25.0%), 41-50 (20.6%) and the least at age ≥ 50 (15.00%) as shown in Table 2. In relation to gender of the patients, the occurrence of K. pneumoniae isolates was higher in females (32.6%) and the male at (15.7%) as shown in Table 3. The occurrence of K. pneumoniae isolates in relation to age and gender of UTI patients were not significant (P > 0.05)
Table 2: Occurrence of Klebsiellapneumoniaerelative to age from urine of patients with suspected Urinary Tract Infection in General Hospital Keffi, Nigeria.
| Age | No. of Samples | No (%) of Klebsiella pneumoniae |
| 11-20 | 32 | 6 (18.7) |
| 21-30 | 81 | 22 (27.1) |
| 31-40 | 48 | 12 (25.0) |
| 41-50 | 29 | 6 (20.66) |
| ≥50 | 20 | 3 (15.0) |
| Total | 210 | 49 (23.3) |
Table 3: Occurrence of Klebsiellapneumoniaerelative to gender from urine of patients with suspected Urinary Tract Infection in General Hospital, Keffi, Nigeria.
| Gender | No. of Samples | No. (%) of Klebsiella pneumoniae |
| Male | 95 | 15(15.7) |
| Female | 104 | 34(32.6) |
| Total | 210 | 49 (12.8) |
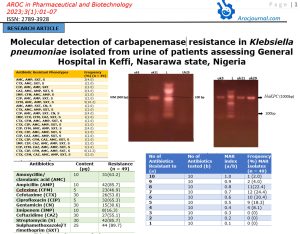
3.3 Antibiotic Resistance
The antibiotic resistance of K. pneumoniae is as shown in Table 4. The K. pneumoniae were more resistant to Sulphamethoxazole/Trimethoprim (89.7%) followed by ampicillin and streptomycin (85.7%) and Ciprofloxacin (65.3%) but less resistant to Cefexime (46.9%), Gentamicin (30.6%) and Imipenem (16.3%).
Table 4: Antibiotic Resistant of Klebsiellapneumoniaeisolated from urine with suspected Urinary Tract Infection in General Hospital, Keffi, Nigeria.
| Antibiotics | Content (µg) | Resistance (n = 49) |
| Amoxycillin/ clavulanic acid (AMC) | 10 | 31(63.2) |
| Ampicillin (AMP) | 10 | 42(85.7) |
| Cefexime (CFM) | 5 | 23(46.9) |
| Cefotaxime (CTX) | 30 | 26(53.0) |
| Ciprofloxacin (CIP) | 5 | 32(65.3) |
| Gentamicin (CN) | 30 | 15(30.6) |
| Imipenem (IMP) | 10 | 8(16.3) |
| Ceftazidime (CAZ) | 30 | 27(55.1) |
| Streptomycin (S) | 30 | 42(85.7) |
| Sulphamethoxazole/Trimethoprim (SXT) | 25 | 44 (89.7) |
3.4 Antibiotic Resistance of Phenotypes
The most common antibiotic resistant phenotypes were CTX, CIP, CFM, AMC, AMP, SXT, CN, S (14.2%) and CTX, CIP, CFM, AMC, AMP, SXT, S (12.2%) as shown in Table 5.
Table 5: Antibiotic Resistant phenotype of Klebsiellapneumoniaeisolated from urine with suspected Urinary Tract infection in General Hospital, Keffi, Nigeria.
| Antibiotic Resistant Phenotypes | Frequency (%) (n = 49) |
| AMC, AMP, SXT, S | 2(4.0) |
| CTX, AMC, SXT, S | 1(2.0) |
| CIP, AMC, AMP, SXT | 1(2.0) |
| CAZ, AMC, AMP, SXT, S | 2(4.0) |
| IMP, CFM, AMC, AMP, S | 1(2.0) |
| CIP, AMC, AMP, SXT, S | 1(2.0) |
| CFM, AMC, AMP, SXT, S | 5(10.2) |
| AMC, AMP, SXT, CN, S | 1(2.0) |
| CTX, AMC, AMP, SXT, S | 1(2.0) |
| CIP, AMC, AMP, SXT, CN, S | 2(4.0) |
| IMP, CTX, CFM, CAZ, SXT, S | 1(2.0) |
| CTX, CFM, AMC, AMP, SXT, S | 1(2.0) |
| CTX, CIP, AMC, AMP, SXT, S | 1(2.0) |
| CIP, CFM, AMC, AMP, SXT, S | 2(4.0) |
| CTX, CIP, AMC, AMP, SXT, S | 1(2.0) |
| CIP, CAZ, AMC, AMP, SXT, S | 1(2.0) |
| CTX, CIP, CFM, CAZ, AMC, SXT, S | 1(2.0) |
| IMP, CIP, CAZ, AMC, AMP, SXT, S | 1(2.0) |
| CIP, CFM, CAZ, AMC, AMP, SXT, S | 1(2.0) |
| CTX, CIP, CFM, AMC, AMP, SXT, S | 6(12.2) |
| CTX, CFM, CAZ, AMC, AMP, SXT, S | 1(2.0) |
| CTX, CIP, CFM, CAZ, AMC, AMP, SXT | 1(2.0) |
| IMP, CTX, CAZ, AMC, AMP, SXT, S | 1(2.0) |
| CTX, CIP, CFM, CAZ, AMC, AMP, SXT, S | 2(4.0) |
| IMP, CIP, CAZ, AMC, AMP, SXT, CN, S | 1(2.0) |
| CTX, CIP, CFM, AMC, AMP, SXT, CN, S | 7(14.2) |
| CRO, CIP, CFM, F, AML, AMP, SXT, CN, S | 2(4.0) |
| IMP, CTX, CIP, CFM, CAZ, AMC, AMP, SXT, CN, S | 1(2.0) |
AMC = Amoxycillin/clavulanic acid, AMP = Ampicillin, CFM = Cefexime, CTX = Cefotaxime, CIP = Ciprofloxacin, CN = Gentamicin, IMP = Imipenem, CAZ= Ceftazidime, S = Streptomycin, SXT = Sulphamethoxazole/Trimethoprim
3.5 Multiple Antibiotic Resistance (MAR) Index
Multiple antibiotic resistance is defined as resistance to more than two antibiotics tested. The MAR indices of the isolates is as shown in Table 6 were > 0.2 with the most common being 0.7(24.4%) and 0.8 (22.4%).
3.6 Categories of Antibiotics
The distribution of the isolates into different categories of antibiotic resistance namely multi-drug resistance (MDR), Extended drug resistance (XDR), Pan drug resistance (PDR) is as shown in Table 7. The order of percentage occurrence was MDR (83.6%) >PDR (16.3) > XDR (0.0%)
Table 6: Multiple Antibiotic Resistance (MAR) Index of Klebsiellapneumoniaeisolated from urine of patients with suspected Urinary Tract Infection in General Hospital, Keffi
| No of Antibiotics Resistant to (a) | No of Antibiotics tested (b) | MAR index (a/b) | Frequency (%) MAR Isolates (n = 49) |
| 10 | 10 | 1.0 | 1 (2.0) |
| 9 | 10 | 0.9 | 2 (4.0) |
| 8 | 10 | 0.8 | 11(22.4) |
| 7 | 10 | 0.7 | 12 (24.4) |
| 6 | 10 | 0.6 | 10 (20.4) |
| 5 | 10 | 0.5 | 9 (18.3) |
| 4 | 10 | 0.4 | 4 (8.1) |
| 3 | 10 | 0.3 | 0 (0) |
| 2 | 10 | 0.2 | 0 (0) |
| 1 | 10 | 0.1 | 0 (0) |
Table 7: Classification of categories of Antibiotics Resistance in Klebsiellapneumoniaefrom urine of patients with suspected urinary tract infection in General Hospital, Keffi, Nigeria
| Categories of Antibiotic Resistance | Frequency (%) (n=49) |
| MDR | 41 (83.6) |
| XDR | 0 (0.0) |
| PDR | 8 (16.3) |
MDR = Multi-drug resistance (non-susceptible to ≥ 1 agent in ≥ 3 antimicrobial): XDR = Entensive drug resistance (non-susceptible to ≥ 1 agent in all but ≤ 2 antimicrobial categories): PDR = Pan drug resistance (non-susceptible to all antimicrobials listed)
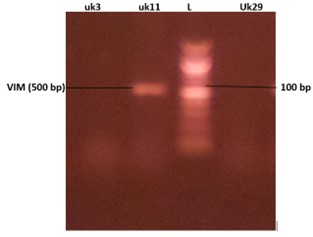
3.7 Molecular Detection of Carbapenem Resistance Gene in K. pneumoniae Isolates
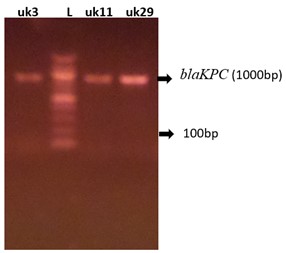
The genotypic detection of carbapenem-resistant genes to Imipenem resistant K. pneumoniae isolates are as shown in Table 4 and figures 1 and 2. The 3 resistant Imipenem resistant K. pneumoniae isolates were blakPC (100%)positive and blaVIM (33.3%) positive.
4.0 Discussion
Carbapenem resistance in K. pneumoniae have been reported to emerge in developing and developed countries [24]. Studies on molecular detection of Carbapenem resistance gene in Imipenem resistant K. pneumoniae isolates in the study location was carried out. The occurrence of K. pneumoniae from urine of patients with suspected Urinary Tract Infection in this study was not new as this finding is similar to studies earlier reported [23][25]. The percentage of occurrence of K. pneumoniae as observed in this study was less than 26.9% and 75.7% as earlier reported by [23][26]. The occurrence of K. pneumoniae in urine of suspected urinary tract infection was an indication that such organism may be responsible for urinary tract infection and this finding is in agreement with the study earlier reported [24, 26] that K. pneumoniae is the most common etiological agent of urinary tract infections.
The occurrence of K. pneumoniae from urine of patients with suspected urinary tract infections in relation to age as observed in this study was high in age 21-30 years and this finding is in agreement with the study earlier reported [27] who reported high prevalence of K. pneumoniae from urine of patients in same age. The high occurrence of K. pneumoniae in age group in this study was an indication the level of hygiene of that age group was low or the level of sexual activity maybe high [28].
The findings in this study also shows the occurrence of K. pneumoniae was high in female than male and this is in agreement with the study earlier described Gonzalez-Padilla et al., [28]. The high prevalence of K. pneumoniae in females compared to may be due to difference in the anatomy of the reproductive system where women had very short urethra, closeness to the anus [23]. The percentage of K. pneumoniae in female in this study was less than 75% reported in (Wadekar & Sathish 2017), less than 69% in females which is higher than 31.0% in males [23] less than 59.3% in females which higher than 40.7% in males [29].
The high resistance of K. pneumoniae isolates to Suphamethoxazole/Trimethopin, Streptomycin, Ampicillin and Ciprofloxacin as observed in this study was not new. This may be due to indiscriminate use of these antibiotics in the study location. The percentage resistance of the isolates to Suphamethoxazole/Trimethopin as observed in this study was higher than 85.8%, and 67.9% earlier reported [29]
The low resistance of the isolates Ceftriaxone, Gentamicin and Imipenem observed in this study was an indication that these antibiotics may not have been abused in the study location. The percentage resistance of the isolates, K. pneumoniae to Gentamicin, Imipenem, was less than 47.8% and 11.2% reported by Akhtar et al., 2016. The low resistance of the K. pneumoniae isolates to antibiotics mentioned justify their use as a common drug of choice for treatment caused by gram negative bacterial infections [23].
The occurrence of Multi-drug resistance (MDR) K. pneumoniae from urine of patients observed in this study was expected and it is in agreement with the study earlier reported [25] that Multi-drug resistance (MDR) K. pneumoniae have been associated with urinary tract infection, thus difficult to be treated. The percentage occurrence of Multi-drug resistance (MDR) K. pneumoniae as observed in this study was less than 50.7% reported by Alhashash et al., [25]
The detection of Carbapenem resistance gene KPC and VIM in Imipenem resistant isolates as observed in this study is in agreement with the study earlier reported [30]. The detection of this gene in the resistant K. pneumoniae isolates was an indication the gene may be responsible for resistance to Imipenem. Although other genes such as SPM, NDM and IPM were not detected in Imipenem resistant K. pneumoniae isolates in this study but have been reported by Ojdana et al. [26] to be responsible for resistance to carbapenem antibiotics. The percentage detection of KPC gene in resistant K. pneumoniae isolates observed in this study was higher than 48.8% reported by previous researcher
5.0 Conclusion
The occurrence of K. pneumoniae isolates from urine of suspected urinary tract infections of patients in this study location was high and antibiotics such as Cefexime, Gentamicin and Imipenem were very effective against the K. pneumoniae isolates. Also, most of the isolates were multi-drug resistance (MDR). In addition, KPC and VIM genes were predominantly detected in imipenem resistant isolates.
Funding: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for profit sectors.
Author’s contributions: All authors contributed to the manuscript and approved the published version
Conflict of interest: The authors declared no conflict of interest.
Acknowledgements: Not Applicable
1. Li, B., et al., Molecular pathogenesis of Klebsiella pneumoniae. Future microbiology, 2014. 9(9): p. 1071-1081.
2. Yao, H., et al., Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. The Lancet Infectious Diseases, 2018. 18(1): p. 25.
3. Roh, K.H., et al., Isolation of a Klebsiella pneumoniae isolate of sequence type 258 producing KPC-2 carbapenemase in Korea. The Korean Journal of Laboratory Medicine, 2011. 31(4): p. 298.
4. Touati, A. and A. Mairi, Epidemiology of carbapenemase-producing Enterobacterales in the Middle East: a systematic review. Expert Review of Anti-infective Therapy, 2020. 18(3): p. 241-250.
5. Yan, J., et al., Multidrug resistance mechanisms of carbapenem resistant Klebsiella pneumoniae strains isolated in Chongqing, China. Annals of laboratory medicine, 2017. 37(5): p. 398-407.
6. Uzoamaka, M., et al., Bacterial etiology of lower respiratory tract infections and their antimicrobial susceptibility. The American Journal of the Medical Sciences, 2017. 354(5): p. 471-475.
7. Egbe, C.A., C. Ndiokwere, and R. Omoregie, Microbiology of lower respiratory tract infections in Benin City, Nigeria. The Malaysian journal of medical sciences: MJMS, 2011. 18(2): p. 27.
8. Marchaim, D., et al., The burden of multidrug-resistant organisms on tertiary hospitals posed by patients with recent stays in long-term acute care facilities. American journal of infection control, 2012. 40(8): p. 760-765.
9. Woldu, M., Klebsiella pneumoniae and its growing concern in healthcare settings. Clin Exp Pharmacol, 2016. 6(1): p. 1-7.
10. Nicolau, D.P., Carbapenems: a potent class of antibiotics. Expert opinion on pharmacotherapy, 2008. 9(1): p. 23-37.
11. Perez, F. and D. Van Duin, Carbapenem-resistant Enterobacteriaceae: a menace to our most vulnerable patients. Cleveland Clinic journal of medicine, 2013. 80(4): p. 225.
12. Coulthurst, S.J., A.M. Barnard, and G.P. Salmond, Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nature Reviews Microbiology, 2005. 3(4): p. 295-306.
13. Remya, P., M. Shanthi, and U. Sekar, Prevalence of blaKPC and its occurrence with other beta-lactamases in Klebsiella pneumoniae. Journal of Laboratory Physicians, 2018. 10(04): p. 387-391.
14. Sah, S.K. and S. Hemalatha, Extended spectrum Beta lactamase (ESBL) Mechanism of antibiotic resistance and Epidemiology. Int J pharmtech Res, 2015. 7(2): p. 303-9.
15. Breurec, S., et al., Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clinical Microbiology and Infection, 2013. 19(4): p. 349-355.
16. Schultsz, C. and S. Geerlings, Plasmid-mediated resistance in Enterobacteriaceae. Drugs, 2012. 72(1): p. 1-16.
17. Giamarellou, H. and G. Poulakou, Multidrug-resistant gram-negative infections. Drugs, 2009. 69(14): p. 1879-1901.
18. Akwa, V., et al., Geographical Perspective of Nasarawa State. Onaivi Printing and Publishing Company Ltd. Keffi, Nasarawa State, Nigeria, 2007: p. 34-35.
19. Cheesbrough, M., District laboratory practice in tropical countries, part 2. 2005: Cambridge university press.
20. Huse, H., et al., Evaluation of oxacillin and cefoxitin disk diffusion and MIC breakpoints established by the clinical and laboratory standards institute for detection of mecA-mediated oxacillin resistance in Staphylococcus schleiferi. Journal of clinical microbiology, 2018. 56(2): p. e01653-17.
21. Osundiya, O., R. Oladele, and O. Oduyebo, Multiple antibiotic resistance (MAR) indices of Pseudomonas and Klebsiella species isolates in Lagos University Teaching Hospital. African Journal of Clinical and Experimental Microbiology, 2013. 14(3): p. 164-168.
22. Harwood, V.J., J. Whitlock, and V. Withington, Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Applied and Environmental Microbiology, 2000. 66(9): p. 3698-3704.
23. Lawal, B., et al., Human Genetic Markers and Structural Prediction of Plasmodium falciparum Multidrug Resistance Gene (pfmdr1) for Ligand Binding in Pregnant Women Attending General Hospital Minna. Journal of Environmental and Public Health, 2018. 2018: p. 3984316.
24. Shahina, Z., et al., A study of antibacterial susceptibility and resistance pattern of E. coli causing urinary tract infection in Chittagong, Bangladesh. Asian J. Biol. Sci, 2011. 4: p. 548-555.
25. Alhashash, F., et al., Multidrug-resistant Escherichia coli bacteremia. Emerging infectious diseases, 2013. 19(10): p. 1699.
26. Ojdana, D., et al., Genetic basis of enzymatic resistance of E. coli to aminoglycosides. Advances in medical sciences, 2018. 63(1): p. 9-13.
27. Tanko, N., Retrospective studies on the prevalence of uropathogens in sokoto metropolis. African Journal of Microbiology Research, 2015. 9(20): p. 1366-1370.
28. Gonzalez-Padilla, M., et al., Gentamicin therapy for sepsis due to carbapenem-resistant and colistin-resistant Klebsiella pneumoniae. Journal of Antimicrobial Chemotherapy, 2015. 70(3): p. 905-913.
29. Akhtar, N., R. Rahman, and S. Sultana, Antimicrobial sensitivity pattern of Escherichia coli causing urinary tract infection in Bangladeshi patients. American Journal of Microbiological Research, 2016. 4(4): p. 122-125.
30. Pezzani, M.D. and S. Antinori, Introduction to urinary tract infections: An overview on epidemiology, risk factors, microbiology and treatment options. Imaging and Intervention in Urinary Tract Infections and Urosepsis, 2018: p. 7-16.