1.0 Introduction
Lactic acid bacteria (LAB) are significant groups of probiotic organisms in fermented food and are generally considered safe (1). Lactic acid bacteria refer to a taxonomically diverse group of Gram-positive bacteria, facultative anaerobic, non-spore-forming, non-motile, and acid-tolerant cocci, coccobacilli, or rods that appear as single cells or forming couples, tetrads, or long chains, with common metabolism and physiology capable of fermenting sugars primarily into lactic acid (2).
LAB is safe microorganisms for both animals and humans. They have a GRAS status (generally recognized as safe) declared by the FDA (USA) as well as the status – QPS (qualified presumption of safety) assigned to them by EFSA (European Food Safety Authority) (EFSA Panel on Biological (3). The lactic acid bacteria (LAB) are isolated from various food matrices and those isolates with better performances and high competitiveness are used as probiotics (4). LAB, such as the Lactobacillus species and yeasts, such as Rhizopus oligosporus, Saccharomyces cerevisiae and the Aspergillus species, are the main producers of fermented rice bran product (5).
Interestingly, the strain mediating lactic acid fermentation of rice can determine some of the properties of the fermented product. For example, Lactobacillus brevis produces gamma-amino butyric acid (GABA) with rice bran, and the fermented rice bran product shows antihypertensive properties (6).
Probiotics are live organisms that confer health benefits on the host when consumed in adequate amounts, by bringing the microbial balance in the system. They are proficient in inhibiting the growth of pathogenic organisms through different mechanisms such as adherence to epithelial cells, modulation of the immune system, and secretion of antimicrobial compounds (7). This proves their ability in the bio preservation of food and can be used as starter culture in the fermentation process under controlled conditions. Therefore, the isolation and characterization of LAB from different traditional fermented foods and products have gained tremendous attention (8).
A potent probiotic isolate must possess certain characteristics like survival and colonizing ability under different environmental conditions (9). The isolates should be able to withstand low pH of gastric juice with resistance to bile salts and also adhere to epithelial cells (10). They should also offer certain health benefits like antimicrobial activity, anticancer activity, and toxin reducing effects, and boosting immune response. Hence, bacteria adhering to suitable surfaces and survival in the gastrointestinal tract should be confirmed by in vitro evaluation prior to using them as probiotics (11). The health benefits of LAB as probiotics such as lowering the risk of diseases, regulation of allergic response, and improving inhabitants of gastrointestinal tract with better immune response have been reported (12).
The contribution of LAB in inhibiting the growth of pathogenic organisms, reducing their toxin secretions, and increasing the nutritional value and other functionality has already been studied (13). Fermentation is a process by which a carbon source is dissimilated by microorganisms yielding energy without net oxidation. The primary end products of microbial fermentation are generally alcohols and organic acids such as lactic acid, acetic acid, and propionic acid (14).
Fermented rice and its products also exert inhibitory effects against pathogenic microorganisms (15). LAB strains make different classes of chemical compounds. Among them, the bacteriocins group is the best-studied one. Bacteriocins are toxic to microbes and are the most promising primary metabolites for developing antibiotic drugs. Bacteriocins are peptides or proteins synthesized by ribosomes, and they inhibit the growth and reproduction of a variety of bacteria. A comprehensive strategy utilizing a One Health approach targeting human, animal and environmental health is crucial to tackling AMR (1).
In spite of the heath benefit associated with this grains, products of fermented rice has not be exploited as an alternative to curb the problem of antibiotic resistant in nations where it is a staple food. Strains of probiotic lactic acid bacteria have been isolated from various sources; however, much research has not been carried out on probiotic lactic acid bacterial species from fermented rice source. This research was determined on isolation and molecularly identifies beneficial lactic acid bacteria from fermented rice water
2.0 Materials and Methods
2.1 Study Area
This study was carried out in Keffi metropolis of Nasarawa state, Nigeria. Keffi is about 58 km from Abuja, the Federal Capita Territory (FCT), and is about 128 km from Lafia, the capital town of Nasarawa state. Keffi is situated on latitude 8°5’N and longitude 7°50’E, and on the altitude of 850 m above the sea level (16).
2.2 Collection of Raw Materials
Rice samples was purchased from Rusau Market in Keffi local government area, Nassarawa State and were transported in sterile sampling bags to the laboratory of Department of Microbiology, Nasarawa State University, Keffi, Nasarawa state, Nigeria for analysis.
2.3 Preparation of Rice Samples
Two hundred grams (200g) of the rice was washed and rinsed thoroughly to remove dirt and dust that may have contaminated the rice during processing, winnowing, transportation and even during storage. Fermentation was carry out by steeping the rice grains with sterile distilled water (rice: water 1:3) for 72 hours (3days) at ambient temperature.
2.4 Microbiological Analysis
2.4.1 Isolation of LAB from fermented rice water
Lactic acid bacteria were isolated from fermented rice water using the pour plate method described by Cheesebrough (2000). Fermented rice water (10 ml) was collected from the steeped rice after shaking well in the Vortex and diluted in sterile saline (0.85%) solution to obtain concentrations 10 0, 10-1, 10-2 and 10-3 dilutions. Aliquots (100 µl) of the prepared dilutions were inoculated on to petri plates in duplicate using spread plate method. The inoculated plates were incubated at 37 oC for 2 days under anaerobic conditions created by Anaerobe gas packs (Oxoid). Isolated bacteria were purified as pure cultures by repeated sub- culturing on media used for primary isolation.
2.4.2 Biochemical Characterization
The bacterial isolates were characterized using colonial morphology, Gram staining, spore forming and biochemical tests. The biochemical tests conducted include catalase, citrate and sugar fermentation profiles. The isolates were identified by comparing their characteristics with those of known taxa using Bergey’s Manual of Determinative Bacteriology (17).
2.5 Molecular identification and characterization of LAB isolates
2.5.1 DNA extraction
Bacteria cells were harvested from 500μl aliquot of bacteria broth culture using a micro centrifuge at 10,000 g for 1min. The residual pellet was resuspended in 300μl of Resuspension Buffer and 2μl of Lysozyme Solution. The mixture was homogenized by inverting several times thereafter incubated at 37 °C for 1 hour. Resuspended cells were recovered by centrifugation and lysed by adding 300μl of Lysis Buffer after which 2μl RNase A and 8μl proteinase K solution were added; followed by incubation at 60 °C for 10mins. The tube was cooled on ice for 5min.300μl binding buffer was added to the mixture and vortexed briefly; the mixture was cooled on ice for 5 mins and thereafter centrifuged at 10,000g for 5 min. The supernatant was transferred directly into the spin column and centrifuged at 10,000g for 1min to trap the DNA. The trapped DNA was washed twice with washing buffer after which it was eluted with 50μl elution buffer into a clean eppendorf tube. Genomic DNA extraction was carried out with column-based JENA Bioscience Bacteria DNA Preparation Kit following manufacturer’s instructions.
2.5.2 Polymerase Chain Reaction (PCR): (16s rRNA Amplification)
Each PCR reaction mixture consisted of 12.5 µl master mix (2x JENA Ruby hot start master mix), 1µl (10pmol) each of forward primer 27F 5’AGA GTT TGA TCM TGG CTC AG3’ and reverse primer 1492R-5’ TAC GGY TAC CTT GTT ACG ACT T 3’, 1µl DNA template and 9.5µl sterile nuclease free water to make up a total reaction volume of 25 µl. PCR amplification was carried out in an Applied Bio-system 2720 Thermo cycler. The mixture was subjected to an initial denaturation at 94°C for 3min; followed by 35 cycles of denaturation at 94°C for 45s , annealing at 55°C for 60s and extension at 72°C for 60 seconds; and a final extension at 72°C for 10mins. The time was finally extended to 10 minutes at 72 °C; it was then chilled at 4 °C in a GEL [17].
2.5.3 Gel Electrophoreses
PCR products were visualized on a 2 % agarose gel containing ethidium bromide in 0.5x Tris-borate buffer (pH 8.0) using blue led trans illuminator (image attached). A molecular ladder marker (Jena Bioscience, 200bp) was run simultaneously to determine the size of the amplicons.
2.5.4 Sequencing
PCR products were purified and sequenced by Sanger sequencing method using AB1 3730XL sequencer and done by Inqaba biotec, Pretoria, South Africa
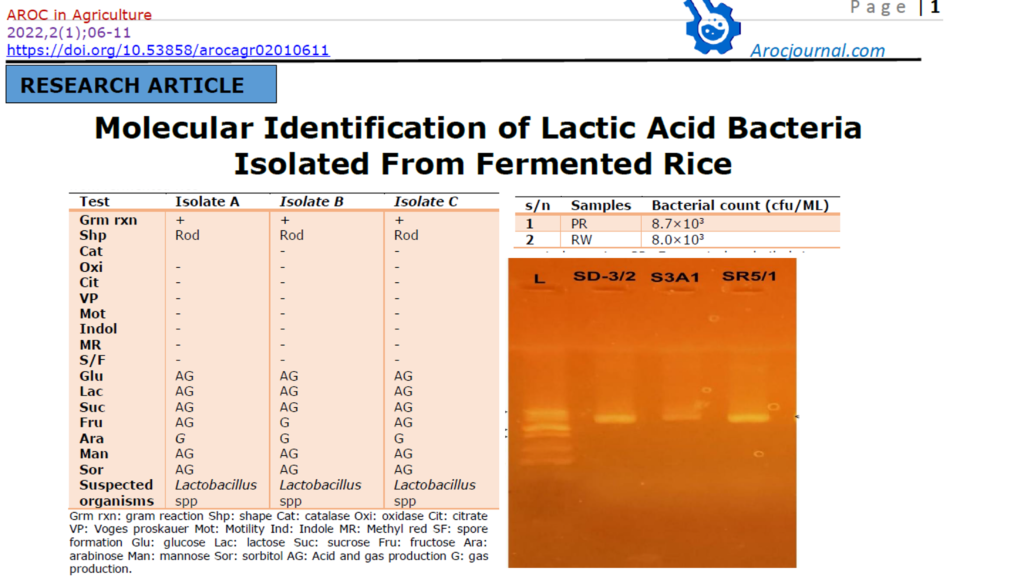
3.1 Bacteria isolate and Biochemical identity of the isolates from fermented rice
The bacterial count of fermented rice water is shown in table 1. The fermented water from raw rice (RW) had the lower bacterial count of 8.0×103 cfu/mL and parboiled rice (PR) has bacterial count 8.0×103 cfu/mL. The microscopic and biochemical tests for identification of bacterial isolates from fermented rice are shown in table 2. Based on the microscopic and biochemical analysis, three (3) suspected strains of lactic acid bacteria were identified from fermented rice.
Table 1 Total aerobic microbial count for fermented rice
| s/n | Samples | Bacterial count (cfu/ML) |
| 1 | PR | 8.7×103 |
| 2 | RW | 8.0×103 |
RW: fermented raw rice, PR: Fermented parboiled rice
Table 2: Microscopic and biochemical tests of bacterial isolates from fermented rice
| Test | Isolate A | Isolate B | Isolate C |
| Grm rxn Shp Cat Oxi Cit VP Mot Indol MR S/F Glu Lac Suc Fru Ara Man Sor Suspected organisms | + Rod – – – – – – – AG AG AG AG G AG AG Lactobacillus spp | + Rod – – – – – – – – AG AG AG G G AG AG Lactobacillus spp | + Rod – – – – – – – – AG AG AG AG G AG AG Lactobacillus spp |
Grm rxn: gram reaction Shp: shape Cat: catalase Oxi: oxidase Cit: citrate VP: Voges proskauer Mot: Motility Ind: Indole MR: Methyl red SF: spore formation Glu: glucose Lac: lactose Suc: sucrose Fru: fructose Ara: arabinose Man: mannose Sor: sorbitol AG: Acid and gas production G: gas production.
3.2 Molecular characteristics of the lactic acid bacteria isolated from fermented rice The result (SD-3/2-SR5/1) show the sequenced amplicons of LAB isolated from fermented rice as the resulting alignment of the concatenated nucleotide (from 5’ 3’ and 3’ 5’) with other known sequences previously blasted in have waves laboratory Ibadan. The gel documented images of the isolated bacterial DNA after electrophoresis appeared at 1500kb which indicated pure isolates. Lane L represents molecular marker (ladder), lane SD-3/2, S3A1 and SR5/1 represents DNA extracted from fermented rice as shown in figure 1
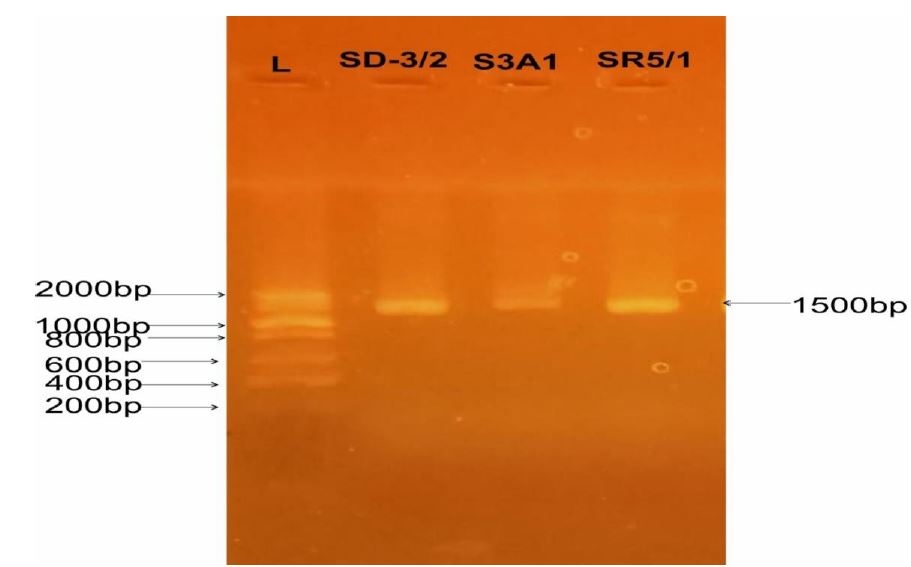
Figure 1: Amplified image of the LAB isolates from Fermented rice. Lane SR5/1; lactiplantibacillus plantarum CIP 103151 lane Sd-3/2; Limosilactobacillus fermentum CIP 102980andlane S3A1; didn’t generate good data.
4.0 Discussion
LABS are safe microorganisms with abilities to produce beneficial compounds (18). With the significant reclassification of LAB in 2020 the scientific name of Lactobacillus plantarum became Lactiplantibacillus plantarum (15). The 16s sequence generated using AB1 3730XL sequencer and forward primer 27F 5’AGA GTT TGA TCM TGG CTC AG3’ and reverse primer 1492R-5’ TAC GGY TAC CTT GTT ACG ACT T 3’, were aligned with sequences in this database and the sequences that produced similar alignment 94.44% assumed Lactiplantibacillus plantarum and95.31% Limosilactobacillus fermentum identity of the isolate. Using blue led trans illuminator. A molecular ladder marker (Jena Bioscience, 2000bp) was run simultaneously to determine the size of the amplicons. Probiotics play a potential alternative to antibiotics in the treatment of inflammatory bowel disease (IBD) (19).
The continuous increase in multiple resistance pathogenic bacteria particularly in the clinical setting has led to the investigation of natural effective alternatives to known antibiotics. Lactic acid bacteria are well known producers of antimicrobial compounds especially bacteriocins which have high antimicrobial activity.
Probiotics have been widely considered for the treatment and prevention of infectious dis- eases because of the various mechanisms In this regard, several studies have investigated the effects of probiotic and prebiotics use on micro biota of other parts of human body (20). Therefore, probiotics are promising in improving skin health by modulating the cutaneous mi- crobiota (21).
By translation, probiotic lactic acid bacteria should be exploited to achieving quality advancement in one health: integrated and unify approach aim at sustainably balance and optimize the health of people, animals and ecosystem. The use of LAB strains in foodomics techniques is highly recommended, as the bacteriocins and volatile compounds in LAB strains could improve the nutritional quality, Since L. plantarum is listed as GRAS (microorganisms and microbial-derived ingredients used in food; generally recognized as safe), its bacteriocins are also considered safe to be use in the food industry as bio preservatives.
5.0 Conclusion
This study has confirmed fermented rice to possess probiotic Lactic acid bacteria. The strains of LA isolated: Limosilactobacillus fermentum and lactiplantibacillus plantarum have been documented to have probiotics potentials and can produce antimicrobial substances. The findings in this study suggest that rice and other locally cultivated grains should be exploited for probiotic organisms that could be useful to man, animals, and the ecosystem.
Author Contributions: This work was carried out in collaboration of all authors. SSM conducted the research and wrote the manuscript. MDM supervised the work. JEO Co-supervised the work. All authors read and approved the final version of the manuscript
Funding: This research received no external funding.
Conflicts of Interest: The authors declare no conflict of interest.
Acknowledgments: The authors are grateful to the technical staff and lecturers of the Department of Microbiology, Faculty of Natural and Applied Sciences, Nasarawa State University Keffi, Nasarawa, Nigeria.
References
1. Raman J, Kim J-S, Choi KR, Eun H, Yang D, Ko Y-J, et al. Application of lactic acid bacteria (LAB) in sustainable agriculture: Advantages and limitations. International Journal of Molecular Sciences 2022;23:7784
2. Khalid K. An overview of lactic acid bacteria. Int J Biosci 2011;1:1-13
3. Binati RL, Salvetti E, Bzducha-Wróbel A, Bašinskienė L, Čižeikienė D, Bolzonella D, et al. Non-conventional yeasts for food and additives production in a circular economy perspective. FEMS Yeast Research 2021;21:foab052
4. Somashekaraiah R, Shruthi B, Deepthi B, Sreenivasa M. Probiotic properties of lactic acid bacteria isolated from neera: a naturally fermenting coconut palm nectar. Frontiers in Microbiology 2019;10:1382
5. Sakamoto N, Tanaka S, Sonomoto K, Nakayama J. 16S rRNA pyrosequencing-based investigation of the bacterial community in nukadoko, a pickling bed of fermented rice bran. International journal of food microbiology 2011;144:352-9
6. Oduguwa OO, Edema MO, Ayeni AO. Physico-chemical and microbiological analyses of fermented corn cob, rice bran and cowpea husk for use in composite rabbit feed. Bioresource technology 2008;99:1816-20
7. Williams NT. Probiotics. American Journal of Health-System Pharmacy 2010;67:449-58
8. Alonso S, Carmen Castro M, Berdasco M, de la Banda IG, Moreno-Ventas X, de Rojas AH. Isolation and partial characterization of lactic acid bacteria from the gut microbiota of marine fishes for potential application as probiotics in aquaculture. Probiotics and antimicrobial proteins 2019;11:569-79
9. Morelli L. In vitro assessment of probiotic bacteria: from survival to functionality. International Dairy Journal 2007;17:1278-83
10. Gupta V, Garg R. Probiotics. Indian journal of medical microbiology 2009;27:202-9
11. Chiang S-S, Pan T-M. Beneficial effects of Lactobacillus paracasei subsp. paracasei NTU 101 and its fermented products. Applied Microbiology and Biotechnology 2012;93:903-16
12. Reuben RC, Roy PC, Sarkar SL, Alam R-U, Jahid IK. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC microbiology 2019;19:1-20
13. Bartkiene E, Zavistanaviciute P, Lele V, Ruzauskas M, Bartkevics V, Bernatoniene J, et al. Lactobacillus plantarum LUHS135 and paracasei LUHS244 as functional starter cultures for the food fermentation industry: Characterisation, mycotoxin-reducing properties, optimisation of biomass growth and sustainable encapsulation by using dairy by-products. Lwt 2018;93:649-58
14. Sieuwerts S. Microbial interactions in the yoghurt consortium: current status and product implications. SOJ Microb Infect Dis 2016;4:1-5
15. Zheng J, Wittouck S, Salvetti E, Franz CM, Harris HM, Mattarelli P, et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. International journal of systematic and evolutionary microbiology 2020;70:2782-858
16. Makut M, Abdulazeez U. A survey of antibiotic-producing actinomycetes in the soil environment of Keffi metropolis, Nasarawa State, Nigeria. J Microbiol Antimicrob 2012;4:74-8
17. Whitman W, Goodfellow M, Kämpfer P, Busse H, Trujillo M, Ludwig W, et al. Bergey’s manual of systematic bacteriology, 2nd edn., vol. 5, parts A and B. Springer-Verlag, New York, NY; 2012.
18. Niu X, Long T, Chen Y. Microbiological assay of a mixture of three lactic acid bacteria metabolites. 亚洲传统医药 2016;11:153-7
19. Gionchetti P, Rizzello F, Lammers KM, Morselli C, Sollazzi L, Davies S, et al. Antibiotics and probiotics in treatment of inflammatory bowel disease. World journal of gastroenterology: WJG 2006;12:3306
20. Wieërs G, Belkhir L, Enaud R, Leclercq S, Philippart de Foy J-M, Dequenne I, et al. How probiotics affect the microbiota. Frontiers in cellular and infection microbiology 2020:454 21. Al-Ghazzewi F, Tester R. Impact of prebiotics and probiotics on skin health. Beneficial microbes 2014;5:99-107