1.0 Introduction.
Pelvic inflammatory disease (PID) is basically a disease of various organs (such as the ovaries, fallopian tubes, uterus and endometrium) located in the upper genital tracts of a female. However, this disease is one of the top three prevalent gynaecological problems, basically associated with female reproductive damages such as; fallopian tubal blockage, endometriosis and oophoritis, which in turn leads to 10% infertility and 0.5% mortality among women in the reproductive ages of 25-34 years [1]. The emergence of this life-threatening disease is basically associated with organisms referred to as urogenital pathogens.
Overtime urogenital pathogens especially members of Enterobacteriaceae such as Escherichia coli, Salmonella sp, Klebsiella sp, Pseudomonas sp, Staphylococcus sp and Proteus sp [2] have been regarded as causative agents of PID in most developing countries such as Nigeria and as such, PID is basically regarded as a polymicrobial infection [3]. However, based on the quest to attain a stable health condition, most female patients misuse and abuse certain existing antibiotics, and this, in turn, has led to the emergence of resistant urogenital pathogens, especially those resistant to multiple classes of antibiotics.
The occurrence of multi drug-resistant urogenital pathogens among females in hospitals and communities have increased hospital stay, treatment failure, morbidity and mortality [4]. However, Enterobacteriaceae such as E. coli and K. pneumoniae is fast becoming the causative agent of most hospital and community infections, based on their ability to exhibit both chromosomally and plasmid-mediated resistance. It is therefore imperative to determine multi drug-resistant Enterobacteriaceae such as E. coli and K. pneumoniae which are fast becoming the urogenital pathogens associated with PID patients in most developing countries. This in turn will enhance the development of new efficient antibiotics and vaccines, in many pharmaceutical industries, which will curb the existence and spread of most multi drug-resistant bacteria.
2.0 Materials and methods
2.1 Description of the study area
The study was conducted in Niger State, Nigeria. Niger State is located in the middle belt zone of the country. It lies between latitude 8˚20′N and 11˚30′N and longitudes 3˚30′E and 7˚20′E [1]. It shares common boundaries with other states namely: Zamfara States to the north; Kaduna State to the north-east; Kebbi State to the north-west, Kogi State to the south; Federal Capital Territory (FCT) and Kwara State to the south-east and south-west respectively. The state covers a land area of about (76,363km2) square km (29,484 square miles) [1]. About 85% of the populace in the state is involved in agriculture, particularly farming and they are majorly rural dwellers, while about 15% are involved in are urban dwellers, involved in white-collar jobs, businesses, crafts and arts.
Source: [5]
The state has three zones, each zone with a distinct climate pattern and a defined agricultural system. Zone A is found in the southern part of the state and it comprises Agaie, Bida, Edati, Katcha, Gbako, Lapai, Lavun and Mokwa Local Government Areas; while zone B comprises of Bosso, Chanchaga, Gurara, Kuta, Paikoro Rafi, Shiroro, Suleja and Tafa Local Government Areas and zone C comprises of Agwara, Borgu, Kontogora, Magama, Mariga, Mashegu, Rijau and Wushishi Local Government Areas [1]. Nine Local Government Areas (comprising of 3Local Government Areas from each zone) were randomly selected for this study.
2.2 Sample size
The number of samples to be collected was calculated using the equation below [6]
Where
n= required sample size
T= Confidence level at 95% (standard value of 1.96)
P= Prevalence rate of bacterial infection in Niger state (9.3%) (Source, Niger State Ministry of Health; [1])
m= Margin of error at 5% (Standard value of 0.05)
n= (1.962x0.093 x 0.907)/0.052=129.6 130
The sample size for each L.G.A was (n) =130 x 3= 390 samples.
1170 samples were collected from 9 general hospitals located in selected local government areas of Niger state (Bida, Suleja and Kontagora).
2.3 Inclusion and exclusion criteria
Female patients within the age of 15-54years diagnosed of PID and are attendees of the selected hospitals were recruited for this study. Female patients above 54years and less than 15 years not diagnosed of PID and who are not attendees of the selected hospitals were excluded from this study.
2.4 Ethical clearance
Ethical clearance for this study was sought from the Niger State Hospital Management board, Research and Ethics Committee.
2.5 Collection and transportation of samples
Endocervical swab: Sterile flexible swab stick was used for the collection of swab from the endocervical region of each patient enrolled for the study [7,8,9]. The swab sticks were removed and submerged into normal saline and were taken to the Microbiology Laboratory of Federal University of Technology, Minna for further analysis [10].
Urine samples: 5ml of fresh urine was collected from each female patient diagnosed of PID into a universal bottle. The urine samples were transported to the Microbiology Laboratory of Federal University of Technology, Minna under cold condition [11]. The urine samples were stored at 4oC for 24 hours for further analysis [11].
2.6 Direct examination
Saline wet preparation was carried out in order to rule out the presence of Trichomonas vaginalis which is characteristically associated with a yellow-green discharge, itching, redness and swelling [4].
2.7 Culture of bacteria
The endocervical swabs and urine samples were cultured and incubated on the following media such as Nutrient agar, MacConkey agar and Salmonella- Shigella agar at 37oC for 24hours for the isolation of Gram negative and Gram positive bacteria. Pure culture of each isolate was obtained by continuous sub-culturing using the streak method. The pure isolates were stored on a nutrient agar slant for further identification and characterization [12].
2.8 Gram staining technique
Smear of the isolate was prepared with a wire loop by emulsifying a colony of the isolate with a drop of distilled water on a clean glass slide free of grease and was used to make a thin smear. The smear was air – dried and was heat fixed. The smear was flooded with crystal violet stain for sixty seconds and was washed with clean running tap water. The slide was tipped off and flooded with lugol’s iodine for sixty seconds and washed with clean water. The smear was decolourized rapidly using alcohol and washed immediately with clean water. The smear was flooded with neutral red stain (safranin) for two minutes, washed with clean water and the back was wiped with clean cotton wool and the smear was allowed to air dry. The dried slides were examined microscopically under oil immersion, using x100 objective lens and the results recorded [12-13].
2.9 Biochemical tests
The bacterial isolates were identified based on the following conventional biochemical tests such as; Coagulase, Oxidase, Catalase, Citrate, Urease, Indole and Triple sugar test [13].
Coagulase test: A drop of distilled water was placed on two separate slides and a colony of the test isolates was emulsified on the two slides to make a thick suspension. A loopful of plasma was added to one of the suspension. Observation for agglutination reaction was done within 10seconds of adding plasma cells [13].
Catalase test: Three milliliters of 3% hydrogen peroxide solution was added into a sterile test tube. A sterile wire loop was used to pick colonies of the test isolates and was immersed in the hydrogen peroxide solution. Observation for bubbles was done immediately and results recorded [13].
Triple sugar iron agar test: The test was performed to determine the ability of bacteria to ferment various carbohydrates such as glucose, lactose and sucrose. Inoculation with the test organism was done by stabbing through the centre of the medium to the bottom of the tube and the test organism was streaked on the surface of the agar slants. The agar slants were incubated at 37oC for 24hours. Observations for colour change of the phenol red indicator to yellow both at the butt and slants (due to the fermentation of either glucose, lactose or sucrose), gas production indicated by bubbles or cracks on the medium and hydrogen sulphide (H2S) production indicated by black pigment coloration was done and recorded [13].
Citrate test: Simmon citrate agar slants were prepared and the surfaces were streaked with isolates and incubated at 37oC for 24hours. Observation for colour change was done and the results were recorded [13].
Indole test: A wire loop of the test organisms was inoculated in the test tubes containing peptone broth at 37oC for 24hours. After 24 hours, 0.5ml of Kovac reagents was added into the test tubes and the solution was thoroughly mixed. Observations for colour change was made and results recorded [13].
Oxidase test: A piece of filter paper was placed in a sterile Petri dish and two drops of freshly prepared oxidase reagent (referred to as tetra-p-diaminechloride) was applied onto the piece of filter paper. A colony of the test organisms was introduced onto the soaked filtered paper. Observation for blue- purple colour within a few seconds was done and the result recorded [13].
2.10 Antimicrobial susceptibility testing of isolates
2.10.1 Preparation of turbidity standard for the inoculums
The McFarland standard was employed in the standardization of the test organisms. Morphologically similar colonies of each test organism was transferred aseptically from an agar plate culture into a tube containing 4 to 5 ml of nutrient broth. The broth was subjected to agitation and was incubated at 37°C until it achieved or exceeded the turbidity of the 0.5 McFarland standard. The turbidity of the actively growing culture in the broth was adjusted with sterile saline or broth to obtain turbidity that was optically comparable to that of the 0.5 McFarland standard [14].
2.10.2 Inoculation of plates
Susceptibility test of the isolates was carried out using Kirby- Bauer disc diffusion method on Mueller-Hinton agar [15]. A sterile cotton swab stick was dipped into the adjusted suspension. The swab stick was rotated several times by pressing it firmly on the inside wall of the tube above the fluid level to remove excess inoculums from the swab stick [12]. The surface of the sterile agar was inoculated by streaking the swab over the entire sterile agar surface. This procedure was repeated by streaking two or more times, rotating the plates approximately 60o each time, to ensure uniform distribution of bacteria on the plates. The inoculated plates was left for 10 minutes to ensure prediffussion of the organisms and to allow excess surface moisture to be absorbed, before the agar plates are impregnated with discs. Each disc was pressed down to ensure complete contact with the agar surface. The plates were inverted and incubated at 37oC for 24h [15].
2.10.3 Reading plates and interpreting results
After 24h hours of incubation, each plate was examined and the diameters of the zones of inhibition were measured, including the diameter of the disc. Zones of inhibition were measured to the nearest whole millimeter using a meter rule [12].
2.11 Screening for antibiotics resistant bacteria isolates
Antibiotics available in the study areas were used for the study. The antibiotic includes: penicillin G (10μg), augmentin (30μg), streptomycin(10μg), ciprofloxacin(5μg), nalidixic acid(30μg), gentamycin (10μg), ofloxacin (5μg), chloramphenicol(10μg) and so on [4].
2.12 Screening for multi drug resistance bacteria
Bacteria isolates resistant to three or more classes of antibiotics according to the clinical laboratory standard institute [15] guidelines were termed multi-drug resistant (MDR) bacteria [16,17].
3.0 Results
3.1 Prevalence of bacteria isolate
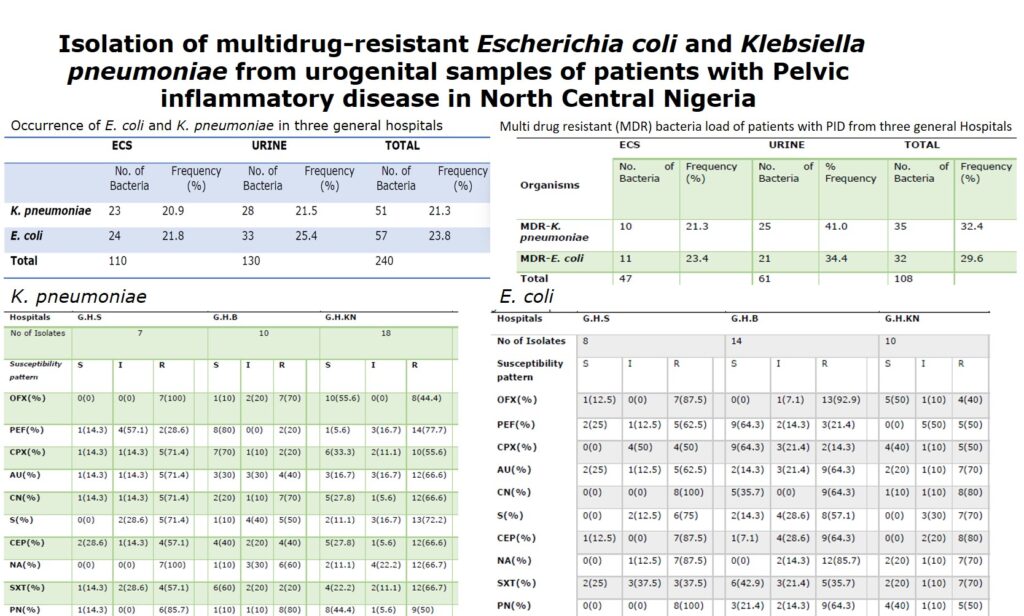
Out of 390 endocervical swabs and urine samples screened, only 240(62%) samples revealed the presence of bacteria (Table 1). The bacteria isolated and identified are indicated in Table 2. It was observed that E. coli was more prominent followed by Klebsiella pneumionae in both endocervical swabs (ECS) and urine samples (Table 2).
Table 1: Prevalence of PID in three general hospitals
| Samples | NSS | NPS | Prevalence (%) |
| Endocervical swap | 390 | 110 | 28.2 |
| Urine | 390 | 130 | 33.3 |
| Total | 240 | 62 |
KEY: NSS= Number of Samples Screened; NPS= Number of Positive Samples
Table 2: Occurrence of E. coli and K. pneumoniae in three general hospitals
| ECS | URINE | TOTAL | ||||
| No. of Bacteria | Frequency (%) | No. of Bacteria | Frequency (%) | No. of Bacteria | Frequency (%) | |
| K. pneumoniae | 23 | 20.9 | 28 | 21.5 | 51 | 21.3 |
| E. coli | 24 | 21.8 | 33 | 25.4 | 57 | 23.8 |
| Total | 110 | 130 | 240 |
KEY: ECS=Endocervical swab
3.2 Prevalence of multidrug resistant K. pneumonia and Escherichia coli
Table 3 shows the frequency of occurrence of multidrug resistant K. pneumoniae 35(46.1%) and Escherichia coli 33(43.4%) from both urine and endocervical samples respectively.
Table 3: Multi drug resistant (MDR) bacteria load of patients with PID from three general Hospitals
| Organisms | ECS | URINE | TOTAL | |||
| No. of Bacteria | Frequency (%) | No. of Bacteria | % Frequency | No. of Bacteria | Frequency (%) | |
| MDR-K. pneumoniae | 10 | 21.3 | 25 | 41.0 | 35 | 32.4 |
| MDR-E. coli | 11 | 23.4 | 21 | 34.4 | 32 | 29.6 |
| Total | 47 | 61 | 108 |
MDR: multidrug-resistant
3.3 Antibiotic Susceptibility Pattern of the isolates
Table 4 and 5 show that more than 50% of the isolated K. pneumoniae and E. coli from each general hospital were resistant to multiple classes of antibiotics
Table 4: Susceptibility Pattern of multidrug resistant Klebsiella pneumoniae in patients with PID from three general hospitals
| Hospitals | G.H.S | G.H.B | G.H.KN | |||||||||
| No of Isolates | 7 | 10 | 18 | |||||||||
| Susceptibility pattern | S | I | R | S | I | R | S | I | R | |||
| OFX(%) | 0(0) | 0(0) | 7(100) | 1(10) | 2(20) | 7(70) | 10(55.6) | 0(0) | 8(44.4) | |||
| PEF(%) | 1(14.3) | 4(57.1) | 2(28.6) | 8(80) | 0(0) | 2(20) | 1(5.6) | 3(16.7) | 14(77.7) | |||
| CPX(%) | 1(14.3) | 1(14.3) | 5(71.4) | 7(70) | 1(10) | 2(20) | 6(33.3) | 2(11.1) | 10(55.6) | |||
| AU(%) | 1(14.3) | 1(14.3) | 5(71.4) | 3(30) | 3(30) | 4(40) | 3(16.7) | 3(16.7) | 12(66.6) | |||
| CN(%) | 1(14.3) | 1(14.3) | 5(71.4) | 2(20) | 1(10) | 7(70) | 5(27.8) | 1(5.6) | 12(66.6) | |||
| S(%) | 0(0) | 2(28.6) | 5(71.4) | 1(10) | 4(40) | 5(50) | 2(11.1) | 3(16.7) | 13(72.2) | |||
| CEP(%) | 2(28.6) | 1(14.3) | 4(57.1) | 4(40) | 2(20) | 4(40) | 5(27.8) | 1(5.6) | 12(66.6) | |||
| NA(%) | 0(0) | 0(0) | 7(100) | 1(10) | 3(30) | 6(60) | 2(11.1) | 4(22.2) | 12(66.7) | |||
| SXT(%) | 1(14.3) | 2(28.6) | 4(57.1) | 6(60) | 2(20) | 2(20) | 4(22.2) | 2(11.1) | 12(66.7) | |||
| PN(%) | 1(14.3) | 0(0) | 6(85.7) | 1(10) | 1(10) | 8(80) | 8(44.4) | 1(5.6) | 9(50) |
Key: OFX: Ofloxacin; PEF: Perfloxacin; CPX: Ciprofloxacin; NA: Nalidixicacid; CN: Gentamicin; ST: Streptomycin; PN: Ampicillin; Cep: Cephalexin; AU: Augmentin; SXT: Sulfamethoxazole; S: Susceptible; I: Intermediate; R: Resistance; G.H.S: General Hospital Suleja; G.H.B: General Hospital Bida; G.H.KN: General Hospital Kontagora
Table 5: Susceptibility pattern of multidrug resistant Escherichia coli in patients with PID from three general hospitals
| Hospitals | G.H.S | G.H.B | G.H.KN | |||||||||
| No of Isolates | 8 | 14 | 10 | |||||||||
| Susceptibility pattern | S | I | R | S | I | R | S | I | R | |||
| OFX(%) | 1(12.5) | 0(0) | 7(87.5) | 0(0) | 1(7.1) | 13(92.9) | 5(50) | 1(10) | 4(40) | |||
| PEF(%) | 2(25) | 1(12.5) | 5(62.5) | 9(64.3) | 2(14.3) | 3(21.4) | 0(0) | 5(50) | 5(50) | |||
| CPX(%) | 0(0) | 4(50) | 4(50) | 9(64.3) | 3(21.4) | 2(14.3) | 4(40) | 1(10) | 5(50) | |||
| AU(%) | 2(25) | 1(12.5) | 5(62.5) | 2(14.3) | 3(21.4) | 9(64.3) | 2(20) | 1(10) | 7(70) | |||
| CN(%) | 0(0) | 0(0) | 8(100) | 5(35.7) | 0(0) | 9(64.3) | 1(10) | 1(10) | 8(80) | |||
| S(%) | 0(0) | 2(12.5) | 6(75) | 2(14.3) | 4(28.6) | 8(57.1) | 0(0) | 3(30) | 7(70) | |||
| CEP(%) | 1(12.5) | 0(0) | 7(87.5) | 1(7.1) | 4(28.6) | 9(64.3) | 0(0) | 2(20) | 8(80) | |||
| NA(%) | 0(0) | 1(12.5) | 7(87.5) | 0(0) | 2(14.3) | 12(85.7) | 2(20) | 1(10) | 7(70) | |||
| SXT(%) | 2(25) | 3(37.5) | 3(37.5) | 6(42.9) | 3(21.4) | 5(35.7) | 2(20) | 1(10) | 7(70) | |||
| PN(%) | 0(0) | 0(0) | 8(100) | 3(21.4) | 2(14.3) | 9(64.3) | 4(40) | 1(10) | 5(50) |
Key: OFX: Ofloxacin; PEF: Perfloxacin; CPX: Ciprofloxacin; NA: Nalidixicacid; CN: Gentamicin; ST: Streptomycin; PN: Ampicillin; Cep: Cephalexin; AU: Augmentin; SXT: Sulfamethoxazole; S: Susceptible; I: Intermediate; R: Resistance; G.H.S: General Hospital Suleja; G.H.B: General Hospital Bida; G.H.KN: General Hospital Kontagora
4.0 Discussion
Pelvic inflammatory disease is basically the disease of female genital organs. It is usually said to occur when bacterial ascends from the vagina to the upper genital tract. This disease occurs as cervicitis, endometritis, oophoritis and salpingitis and in severe cases it can result to infertility [9,18].
This study reveals that 240(62%) of the samples collected from PID patients were positive for bacterial growth. This is based on the silent spread of bacterial organisms to the upper genital tract which results to high degree of damages such as miscarriage, preterm labor and ectopic pregnancy in the infected females [19]. These therefore lead to infertility among the female population. This is in agreement with the findings of [8,19] who reported that higher rates of bacterial infections such as 60%, 57% and 30% in Africa, Asia and Indian respectively.
The high occurrence of E. coli 57(23.8%) revealed in this study, could be based on the fact that E. coli predominantly colonize the gastrointestinal tract and as such is the main causative agent of urinary tract infections, and this frequently exposes this organism to the vagina due to its proximity to the periurethral openings and the perianal areas. This results is in agreement with the findings of [20], who revealed that majority of the organisms isolated from patients with urogenital infections are E. coli.
Furthermore, the highest occurrence of multidrug resistant K. pneumoniae 35(32.4%) in this study, could be attributed to the fact that K. pneumoniae is the predominant organism mostly associated with community and hospital infections respectively. This result also confines to the study of [21], who revealed that 58 bacteria isolates associated with urogenital infections, were multi drug resistant Klebsiella pneumoniae.
High resistance of above 70% exhibited by Klebsiella pneumoniae and Escherichia coli to Ofloxacin, Nalixidic acid, Ampicillin, Gentamicin, Perfloxacin, Streptomycin and Cephalexin could be attributed to the fact that most of these bacterial isolates exhibit high horizontal transfer of genetic materials such as transposons, plasmids which codes for resistance to many classes of antibiotics, which in turn brings about treatment failure with most antibiotics. This result agrees with the findings of [22], who reported that high multidrug resistant isolates were resistant to all antibiotics.
Collectively, this study basically revealed high resistance profile of multidrug resistant Klebsiella pneumoniae and E. coli associated with pelvic inflammatory disease (PID). However, these isolated multidrug resistant organisms are involved in the rapid dissemination of resistant genes among the intra and inter species in the study area, and as such, have led to prolonged hospital stay, high morbidity and high mortality among reproductive age women with pelvic inflammatory disease (PID). Therefore, it is necessary that the government enforce certain measures such as; constant awareness on the misuse of antibiotics and laws against self-medication and illegal purchase of drugs across the counter to help curb the menace associated with pelvic inflammatory disease that have resulted from resistant organisms.
5.0 Conclusion
The results of this study confirmed a high presence of multidrug-resistant Klebsiella pneumoniae and Escherichia coli in pelvic inflammatory disease (PID) patients attending tertiary hospitals in North Central Nigeria, hence there is a need for public health workers, to create awareness on the misuse of antibiotics, to prevent and curtail treatment failure due to antibiotic resistance.
Authors’ Contributions: The work was conducted in collaboration of all authors. All authors read and approved the final version of the manuscript.
Ethical Approval: Ethical clearance for this study was obtained from the Niger State Hospital Management Board, Research and Ethics Committee.
Informed consent: Informed consent was obtained from all individual participants included in the study
Competing Interests: The authors have no conflict of interest.
Funding: No supportive funds were received from external organization.
References
[1]. Usman, M. (2016). Prevalence of polymicrobial pelvic inflammatory infection among women attending three general hospitals in Niger state, Nigeria. A Published Thesis presented to the Department of Microbiology, Federal University of Technology Minna, Niger State, Pp. 33-41.
[2]. Meštrović, T. (2017). Pelvic Inflammatory Disease Etiology. Retrieved May 23rd, 2018 from https://www.news-medical.net/medical
[3]. Centers for Disease Control and Prevention (2015). Sexually transmitted diseasestreatment guidelines. Pelvic inflammatorydisease (PID). Retrieved May 25th, 2018 from, http://www.cdc.gov/std/tg2015/pid.htm
[4]. Spencer,T. H.I., Umeh,P. O., Irokanulo,E., Baba,M. M., Spencer, B. B., Umar, A. I., Ardzard, S.A., Oderinde, S. & Onoja O. (2014). Bacterial Isolates Associated With Pelvic Inflammatory Disease Among Female Patients Attending Some Hospitals in Abuja, Nigeria.African Journal of Infectious Disease, 8(1), 9-13.
[5]. Niger State Bureau of Stastistics (2010). Themap of Niger State.Retrieved May 25th, 2018 from, http://www.ask.com.
[6]. Cochran, W.G. (1977). Sampling techniques.3rd Edn. New York: John Wiley & Sons, New York, Pp 220
[7]. Einwalter,L.A., Ritchie, J.M., Ault, K.A. & Smith, E.M. (2005). Gonorrhea and Chlamydia infection among women visiting family planning clinic: racial variation in prevalence and predictors. Perspective Sex Reproductive Health, 37(3), 135-140.
[8]. Pachori, R. & Kulkarni, N. (2016). Epidermiological markers in pelvic inflammatory disease (PID) among the women of reproductive age group. European Journal of Biomedical and Pharmaceutical Science, 3(2),193-196.
[9]. Oseni, T.I.A. & Odewale, M.A. (2017). Socioeconomic status of parents and the occurrence of pelvic inflammatory disease among undergraduates attending Irrua Specialist Teaching Hospital, Irrua, Edo State, Nigeria. Nigerian Postgraduate Medical Journal, 24, 2, 114-120.
[10]. Enwa, F. O., Iyamu, M. I., Eboigbe, C. I. & Esimone, C.O. (2015). Prevalence of resistant strains of Streptococcus pneumoniae to Oxacillin, Ofloxacin and Rifampicin in Abraka South-South, Nigeria. Global Journal of Medical Research: Microbiology and Pathology,15 (4),1.
[11]. Hunter,K.F., Voaklander, D., Hsu, Z.Y. & Moore, K. (2013).Lower urinary tract symptoms and falls risk among older women receiving home support: a prospective cohort study.BMC Geriatrics,13,46.
[12]. Kolo, S. (2016). Prevalence of Bacteriological and parasitic infections in HIV children (2-16 years) in Niger State. A Published Thesis presented to the Department of Microbiology, Federal University of Technology Minna, Niger State, Pp. 58-79.
[13]. Cheesbrough, M. (2010). District Laboratory Practicein Tropical Countries, Part 2, 2nd Edition update. United Kingdom: Cambridge University Press, Cambridge, Pp 107-114.
[14]. Lalitha, M.K. (2004). Manual on Antimicrobial Susceptibilty Testing. Retrieved May 10th 2014. From www.ijmm.org/documents/antimicrobial.doc.
[15]. Clinical and Laboratory Standards Institute, (2014). Performance Standards for Antimicrobial Susceptibility Testing. 22nd Edn., Clinical and Laboratory Standards Institute, USA., ISBN: 9781562388973, Pages: 226.
[16]. Magiorakos, A.P., Srinivasan, A., Carey, R.B., Carmeli, Y., Falagas, M.E. & Giske, C.G., (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology Infections, 18,268-81.
[17]. Iliyasu, G., Habib, A.G.,Mohammed, A.B. & Borodo, M.M. (2015). Epidemiology and clinical outcomes of community acquired pneumococcal infection in North-West Nigeria. Sub- Saharan Afr J Med,2,79-84.
[18]. Ahmed, S., Parvin, S., Shaha, D., Begum, P., Sanjowal, L., Hassan, M. K. & Mohammad Arif, K. (2017). Clinical Profile of Pelvic Inflammatory Disease (PID). Faridpur Medical College Journal, 12(1), 25-30.
[19]. Naaz,F., Khan, N. & Mastan, A. (2016). Risk factors of pelvic inflammatory disease: A prospective study. Int. Journal of Herbal Medicine, 4(4),129-133.
[20]. Erdem, I., Ali, R.K., Ardic,E., Omar,S.E., Mutlu, R., Topkaya, A.E. (2018).Community-acquired Lower Urinary Tract Infections: Etiology, Antimicrobial Resistance, and Treatment Results in Female Patients. Journal of Global Infectious Disease, 10(3), 129–132.
[21]. Metri, B. C., Jyothi, P. & Peerapur B. V. (2012). Detection of ESBL in E. coli and K. pneumoniae isolated from urinary tract infection in India. Indian J Nephrology, 22(5), 401–402.
[22]. Iseghohi, F. (2016). Prevalence and molecular characterization of extended spectrum betalactamase gene from Escherichia coli isolated from four hospitals in Minna, Niger state.A Published Thesis presented to the Department of Microbiology, Federal University of Technology Minna, Niger State, Pp. 108-112