1.0 Introduction
Xanthomonas campestris gram-negative bacteria, having a single polar flagellum [1], plant-associated bacteria that is generally pathogenic for plants belonging to the family Brassicaceae [2]. They cause huge problems in areas with a warm and wet climate [3]. Machinery and insects damage plants creating wound entry points for Xanthomonas bacteria [4]. The bacteria enter plant leaves through breathing pores (stomata) and water releasing pores (hydathodes) in the leaf surface. Once inside the plant, the bacteria reproduce and move around using the plant’s water transportation system (xylem) [5]. Xanthomonas bacteria feed on nutrients released from plant cells as they become leaky and start to die. They cause black rot disease where leaves become yellow and sticky, then wilt and die [5]. The bacteria are the causal agents of an assortment of persistent and periodic plant diseases [3], most strains are relatively common phytopathogenic bacteria which mainly infect crucifers and cause black rot which has high agronomic and economic importance-particularly in regions of subsistence agriculture and those with poor crop management resources [3]. X. campestris, is the most widely used for the industrial production of xanthan gum-based on its high yield and the high-quality product suitable for many applications [6]. It is strictly aerobic bacteria, and the rate of oxygen transmission affects xanthan gum production [7].
Xanthan gum is produced by Xanthomonas campestris [1,8. It derived its name from the strain of bacteria used during the fermentation process [9]. It has important physicochemical properties including high shear stability, pseudo-plastic features and stability on a wide variety of temperatures and pH values [10]. It’s already a widely used product in the market when compared to the other polysaccharides of plant and microbial origin and it has already been accepted by the US food and drug administration in 1969 and received European Union approval under the E-number 415 in 1980 [7]. Because of its unique rheological behaviour [11], it is one of the major microbial polysaccharides actually employed in many industrial processes [6]. It is tasteless and does not affect the taste of other food ingredients [12]
It has excellent solubility and stability under both acidic and alkaline conditions [13]. Despite its wide application, some problems in production persist [14].Commercially available xanthan is relatively expensive, since it is manufactured from glucose or sucrose [11]. In recent years, considerable research has been carried out using agro-industrial wastes, which are renewable and abundantly available to produce value-added products [5]. The present study was set out for molecular identification of Xanthomonas campestris from leaves of four different plants with indications of dark rot spot and evaluation their xanthan gum production capacity
2.0 Materials and methods
2.1 Description of the study area
The study area was in Keffi, Nasarawa state, Nigeria. Keffi is an historic town located in Nasarawa state, North Central Nigeria between latitudes 80 511 and 80 531 North of the equator and longitudes 70 50’ and 70 51’ East of the Greenwich meridian. Keffi is located about 128km away from Lafia, the Nasarawa state capital and about 57km away from Abuja, the Federal Capital Territory of Nigeria. Keffi is the smallest L.G.A in the whole of Nasarawa state with a total land area of approximately 140km2 [15]
2.2 Media
Media including Nutrient agar (NA), Yeast malt broth (YM) medium, Yeast Carbonate Dextrose Agar (YCDA), Starch agar, Simmon’s citrate agar, Urea broth, Tryptone broth, Luria Bertani (LB) were of analytical grade and were obtained from reputable company
2.3 Sample collection
Hundred (100) plants leaves, twenty-five each for mango, rice, mint plant and pepper, showing black rot symptoms were collected in paper bags each bag [16], within Keffi metropolis and transported to Nasarawa State University Keffi, department of microbiology laboratory for analysis.
2.4 Isolation and screening of xanthan producing bacteria
The leaf samples were cut into small pieces, soaked in 5 ml sterilized water of Yeast extract broth and was incubated for 24 hrs as described by Tatjana et al. [16], and Kassim, [13]. The resultant broth was streaked onto Yeast malt extract agar plate (3 g/l yeast extract, 3 g/l malt extract, 10 g/l glucose, 5 g/l peptone and 20 g/l agar) and the plates was incubated at 30°C for 48 hrs and examined. Isolated colonies were further streaked on Yeast Carbonate Dextrose Agar (YCDA) plates, and incubated at 30°C for 48 hrs. The bacteria with yellow mucoid colonies were selected for further analysis [13].
2.5 Cultural, morphological and biochemical characteristics Identification of the isolate
The colonies were purified on nutrient agar plates for observation, colonies were observed for pigmentation, size and shape as described by Ogolla and Neema [3]. Gram staining were used to study the morphological characteristics and gram reaction as described previously [17-20]. Various biochemical tests were performed for the identification of the isolates like, methyl red test, indole production test, urease test, citrate utilization test, starch hydrolysis, milk Proteolysis, gelatin liquefaction and tween 80-hydrolysis test [3].
2.6 Molecular Identification
2.6.1 DNA extraction
Five milliliters of an overnight broth culture of the bacterial isolate in Luria Bertani (LB) broth were spun at 14000rpm for 3 min. The cells were re-suspended in 500µl of normal saline and heated at 950C for 20 min. The heated bacterial suspension was cooled on ice and spun for 3 min at 14000rpm. The supernatant containing the DNA was transferred to a 1.5ml microcentrifuge tube and stored at -20oC for other downstream reactions as earlier described by Abimiku et al., [21].
2.6.2 DNA quantification
The extracted genomic DNA was quantified using the Nanodrop 1000 spectrophotometer. The software of the equipment was lunched by double clicking on the Nanodrop icon. The equipment was initialized with 2µl of sterile distilled water and blanked using normal saline. Two microliters of the extracted DNA were loaded onto the lower pedestal, the upper pedestal was brought down to contact the extracted DNA on the lower pedestal. The DNA concentration was measured by clicking on the “measure” button [21].
2.6.3 16S rRNA Amplification
The 16s rRNA region of the rRNA genes of the isolates were amplified using the 27F: 5′-AGAGTTTGATCMTGGCTCAG-3′ and 1492R: 5′-CGGTTACCTTGTTACGACTT-3′ primers on ABI 9700 Applied Biosystems thermal cycler at a final volume of 25 microlitres for 35 cycles. The PCR mix included: the X2 Dream taq Master mix supplied by Inqaba, South Africa (Taq polymerase, DNTPs, MgCl), the primers at a concentration of 0.4M and the extracted DNA as template. The PCR conditions were as follows: Initial denaturation, 95°C for 5 minutes; denaturation, 95°C for 30 seconds; annealing, 52°C for 30 seconds; extension, 72ºC for 30 seconds for 35 cycles and final extension, 72°C for 5 minutes [22-23.
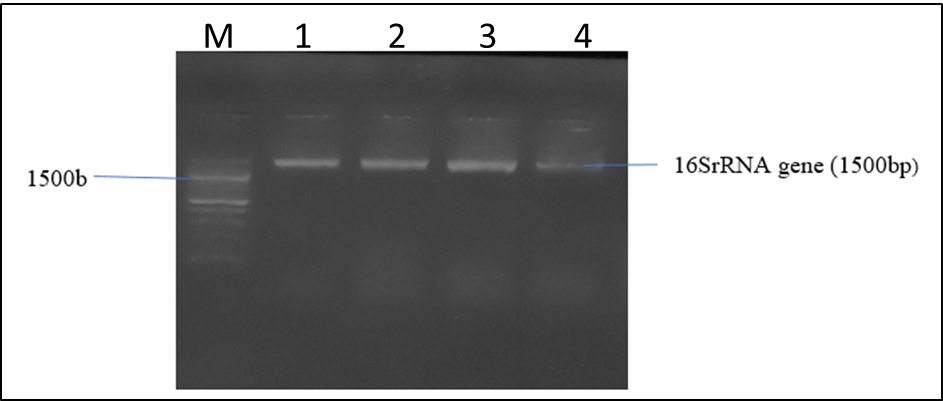
2.6.4 Agarose Gel Electrophoresis of the 16SrRNA gene from the bacteria isolates
The agarose gel electrophoretic separation of digested 16S rRNA gene was carried out as described by Abimiku et al., [21], 8µl of PCR products stained with ethidium bromide was loaded into 1.0% (wt/vol) agarose gel wells with a molecular marker run concurrently at 120 V for 30 min. The DNA bands were visualized and photographed under UV light 595nm.
2.7 Fermentation medium for xanthan production
The following medium was used [Sugar cane molasses, KH2PO4 (5g/l), MgSO4. 7H2O (0.2g/l), citric acid (2g/l), FeCl3.6H2O (0.002 g/l), CaCO3 (0.02g/l), Glutamate (2g/l)] [9]. The medium was sterilized for 20 min at 121°C and medium initial pH was adjusted to 7 [13]. Fermentation was carried out in 250 ml Erlenmeyer flask, each of which contained 100 mL of the sterile production medium. The medium was inoculated with 5 (v/v%) of the inoculum and incubated at 37°C for 72 hrs at 200 rpm. The different isolates were inoculated in these media and after incubation the viscosity of the broth was measured. Isolates giving the highest viscosity was further used for optimization studies [24].
3.0 Results and Discussion
3.1 Cultural, morphological and biochemical identification of the isolates The Xanthomonas sp. strains, plant pathogens, were isolated from the infected leaves of different plants. Primary identification of the Xanthomonas sp. strains were conducted according to pigment production. Several species of bacteria contained identical pigments [25] and X. campestris produces yellow pigment called xanthomonadin [26]. Xanthomonas sp. strains were formed yellow pigmented slimy or mucoid colonies on YMA and NA plates. The cultural, morphological and biochemical characteristics of Xanthomonas species isolated from twelve infected plant leaves and as given in table 1 shows that the organisms appear yellowish in color, gram negative, rod shape and biochemical reactions namely: Starch hydrolysis: positive, methyl red: negative, gelatin liquefaction: positive, Milk proteolysis: positive, Urease production: positive, citrate utilization: negative, indole production: negative (table 1)
Table 1: Cultural, morphological and biochemical identification of the isolates
| Morphology | Biochemical characteristics | |||||||||||
| Isola te | Colony appearance | Gram staining | Shape | Sh | Gl | Mp | Up | Th | Cu | MR | Ip | Inference |
| X1 | Nt_Transp_Yel | – | Rod | + | + | + | + | + | – | – | – | Xanthomonas sp. |
| X2 | Nt_Transp_Yel | – | Rod | + | – | + | + | + | – | + | – | Xanthomonas sp. |
| X3 | Nt_Transp_Yel | – | Rod | + | + | + | + | + | – | – | – | Xanthomonas sp. |
| X4 | Nt_Transp_Yel | – | Rod | + | + | + | + | + | – | – | – | Xanthomonas sp. |
| X5 | Nt_Transp_Yel | – | Rod | + | + | + | – | + | – | – | + | Xanthomonas sp. |
| X6 | Nt_Transp_Yel | – | Rod | + | + | + | + | + | – | – | – | Xanthomonas sp. |
| X7 | Nt_Transp_Yel | – | Rod | + | + | – | + | + | – | – | – | Xanthomonas sp. |
| X8 | Nt_Transp_Yel | – | Rod | + | + | + | – | + | + | – | – | Xanthomonas sp. |
| X9 | Nt_Transp_Yel | – | Rod | + | + | + | + | + | – | – | – | Xanthomonas sp. |
| X10 | Nt_Transp_Yel | – | Rod | + | + | + | + | + | – | – | – | Xanthomonas sp. |
| X11 | Nt_Transp_Yel | – | Rod | + | + | + | + | + | – | – | – | Xanthomonas sp. |
| X12 | Nt_Transp_Yel | – | Rod | + | + | + | + | + | – | – | – | Xanthomonas sp. |
Key: X1 – X12 = Infected leaves; (+) = Positive, (-) = Negative, Sh = Starch hydrolysis, Gl = Gelatin liquefaction, Mp = Milk proteolysis, Up = Urease production, Th = Tween 80 hydrolysis, Not transparent Yellowish= Nt_Transp_Yel, Cu = Citrate utilization, MR = Methyl red, Ip = Indole production.
3.2 Occurrence of Xanthomonas species from infected leaves
The occurrence of Xanthomonas species is as given in Table 4.2. Where the highest occurrence was observed from mint plant (24%) followed by mango leave (12.0%), 4.0% for rice and 8.0% for pepper as shown in Table 2.
Table 2: Occurrence of Xanthomonas species from infected leaves
| Infected leaves | Number sample | No. (%) isolated |
| Mint plants | 25 | 6(24.0) |
| Mango | 25 | 3(12.0) |
| Rice | 25 | 1(4.0) |
| Pepper | 25 | 2(8.0) |
3.3 Screening for xanthan gum production.
It was reported that the presence of glutamate, aspartate, proline, hydroxyproline, threonine and alanine stimulated xanthan gum production [27]. Also, Murad et al. [28] found that several amino acids (cysteine, histidine, glycine and serine) were suitable for xanthan gum production. To produce xanthan gum, X. campestris needs several nutrients, including micronutrients (e.g. K, Fe, P, Mg, S and Ca salts) and macronutrients (C and N) [29]. The CFP was considered as an enhancer of xanthan gum production because of including these amino acids and minerals or salts.
The screening for Xanthan gum product is as shown in Table 4.3. Isolates from plants varying ability in Xanthan gum production coded with (Xh1 – Xh6) for mint plant, (Xm1 – xm3) for mango, (Xr1) for rice and (Xp1 – Xp2) for pepper. Where Xh1 produced (0.10g/l), Xh2 produce (0.8g/l), Xh5 produced (0.9g/l) and Xm2 produced (0.8g/l), as shown in Table 4.3, Xh1 (0.10g/l) and Xm2 (0.8g/l), were further used for production (Table 3). Similar results have been obtained in previous studies [30-31]
Table 3: Screening for xanthan gum production by isolates
| Isolates codes | Xanthan gum (g/l) | |
| Xh1 | 0.10 ± 0.02 | |
| Xh2 | 0.8 ± 0.01 | |
| Xh3 | 0.0 ± 0.00 | |
| Xh4 | 0.0 ± 0.00 | |
| Xh5 | 0.9 ± 0.00 | |
| Xh6 | 0.0 ± 0.00 | |
| Xm1 | 0.0 ± 0.00 | |
| Xm2 | 0.8 ± 0.03 | |
| Xm3 | 0.0 ± 0.00 | |
| Xr1 | 0.0 ± 0.00 | |
| Xp1 | 0.0 ± 0.00 | |
| Xp2 | 0.0 ± 0.00 |
Key: Xh1 – Xh6 = Mint plants, Xm1 – Xm3 = Mango leaves, Xr = Rice leaves, Xp1 – Xp2 = Pepper leaves
3.4 Molecular identification of the Xanthomonas species
The molecular identification of the bacterial isolates is shown in Figure 1. Agarose gel electrophoresis revealed different molecular of the Xanthomonas species isolates with base pair.
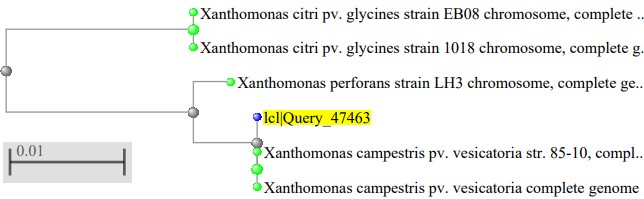
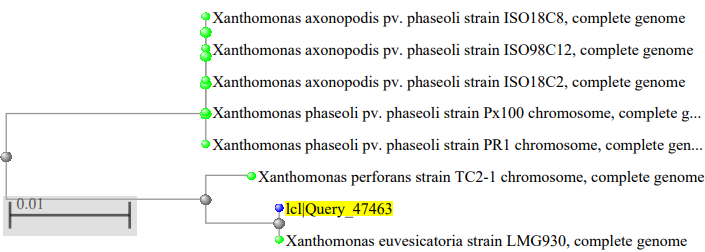
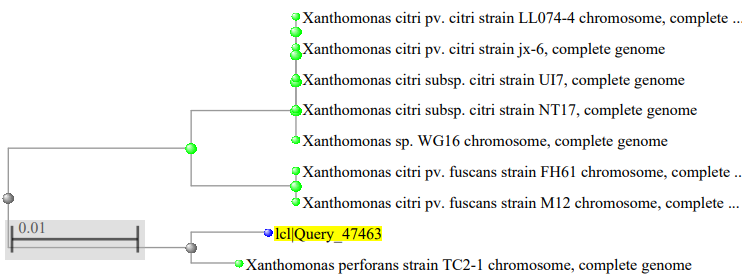
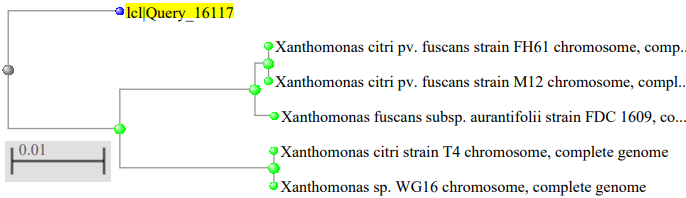
The phylogenetic tree of the Xanthomonas species isolated as showed in figure 2, where the Xanthomonas species isolated from mango were highly related to Xanthomonas campestris pv. vesicatoria strain 85–10, figure 3 showed the Xanthomonas species isolated from pepper were highly related to Xanthomonas euvesicatoria pv. alfalfa strain CFBP3836, figure 4 showed the Xanthomonas species isolated from mint plant were highly related to Xanthomonas perforans strain 91-118 and figure 5 showed the Xanthomonas species isolated from rice were highly related to Xanthomonas citri pv. fuscans strain M12.
Conclusion
Conclusively,Xanthomonas species were isolated from plants leaves in Keffi and they were able to produce xanthan gum. The study confirmed high xanthan gum production capacity of Xanthomonas campestris from dark rot spot containing mint leaves and should be considered during local and industrial production of the xanthan gum.
Author’s contributions: All author participated in study design, methodology, data collection, and original draft writing and manuscript preparation. All authors read and approved the final version of the manuscript
Conflict of interest statement: The authors hereby declare that there is no conflict of interest from any of them.
Funding: This work received no external funding
Institutional Review Board Statement: Not Applicable.
Informed Consent Statement: Not Applicable
References
- Akpan, E.U., Enyi, G.C. and Nasr, G.G. (2020). Enhancing the performance of xanthan gum in water-based mud systems using an environmentally friendly biopolymer. Journal of Petroleum Exploration and Production Technology. 10(1): 1933 – 1948.
- Lopes, D.M.B., Vinicius, L.L., Barbara, M.S., Marco, A.D.S.C.F., Egon, S. and Luiz, G.L. (2015). Xanthan gum: properties, production conditions, quality and economic prospective. Journal of Food and Nutrition Research. 54(3): 185 – 194.
- Ogolla, F.O. and Neema, D.B. (2019). Cultural, morphological and biochemical identification of Xanthomonas Spp. the causative agent of bacteria leaf spot in tomatoes in Wanguru, Mwea, Kirinyaga County, Kenya. International Journal of Research and Innovation in Applied Science (IJRIAS). 4(4): 44 – 49.
- Gazdik, F., Magnus, S., Roberts, S.J., Baranski, R., Cechova, J., Pokluda, R., Eichmeier, A., Grzebelus, D. and Baranek, M. (2021). Persistence of Xanthomonas campestris pv. campestris in field soil in central Europe. Microorganisms. 591(9): 1 – 16.
- Narvekar, S., Thilakan, A., Bhangale, M., Yeri, A., Pawar, V., Kasar, N., Shukla, P. and Trivedi, M. (2019). Isolation, identification and invitro antibiotics sensitivity pattern of local isolates of Xanthomonas followed by production of xanthan gum using agro-industrial waste. JETIR. 6(5): 181 – 188.
- Elella, M.H.A., Goda, E.S., Gab-Allah, M.A., Hong, S.E., Pandite, B., Lee, S., Gamal, H., Rehmanh, A. and Yoon, K.R. (2021). Xanthan gum-derived materials for applications in environment and eco-friendly materials: A review. Journal of Environmental Chemical Engineering. 9(1) 1 – 31.
- Dai, X., Gao, G., Mengmeng, W., Weiying, W., Jianmei, Q., Guoqiang, L. and Ting, M. (2018). Construction and application of a Xanthomonas campestris CGMCC15155 strain that produces white xanthan gum. Microbiology Open. 10: 1 – 9.
- Benkadri, S., Salvador, A., Sanz, T. and Zidoune, M.N. (2021). Optimization of xanthan and locust bean gum in a gluten-free infant biscuit based on rice-chickpea flour using response surface methodology. Food. 10(12): 1 – 12.
- Gomashe, A.V., Dharmik, P.G. and Fuke, P.S. (2013). Optimization and production of xanthan gum by Xanthomonas campestris NRRL-B-1449 from sugar beet molasses. The International Journal of Engineering and Sciences (IJES). 5(2): 52 – 55.
- Tahereh, G., Mohammad, R.S. and Saman, H. (2016). Optimization of xanthan gum production from grape juice concentrate using plackett-burman design and response surface methodology. Applied Food Biotechnology. 3(1): 15 – 23.
- Mabrouk, M.E.M., El-Ahwany, A.M.D., Beliah, M.M.B. and Sabry, S.A. (2013). Xanthan production by a novel mutant strain of Xanthomonas campestris: application of statistical design for optimization of process parameters. Life Science Journal. 10(1): 1660 – 1667.
- Das, B., Sujata, P. and Kumar, S.H. (2021). A review on bio-polymers derived from animal sources with special reference to their potential applications. Journal of Drug Delivery and Therapeutics. 11(2): 209 – 223.
- Kassim, M.B.I. (2011). Production and characterization of the polysaccharide “xanthan gum” by a local isolate of the bacterium Xanthomonas campestris. African Journal of Biotechnology. 74(10): 16924 – 16928.
- Ozdal, M. and Kurbanoglu, E.B. (2018). Valorisation of chicken feathers for xanthan gum production using Xanthomonas campestris MO-03. Journal of Genetic Engineering and Biotechnology. 16: 259 – 263.
- Yakubu, I.O. (2013). Geography of Keffi, Nasarawa state, Nigeria. www.academia.edu/13014831 (published online in 2013). Pg 1 – 11.
- Tatjana, P., Dragana, J., Mira, S., Milovanović, P., Dolovac, N., Poštić, D and Stanković, S. (2013). Phenotypic and genotypic characterization of Xanthomonas campestris strains isolated from cabbage, kale and broccoli. Arch. Biol. Sci., Belgrade, 65 (2), 585 – 593.
- Lawal, B., Shittu, O.K., Oibiokpa, F.I., Mohammed, H., Umar, S.I. and Haruna, G.M., 2016. Antimicrobial evaluation, acute and sub-acute toxicity studies of Allium sativum. Journal of Acute Disease, 5(4), pp.296-301.
- Yusuf, A.A., Lawal, B., Yusuf, M.A., Adejoke, A.O., Raji, F.H. and Wenawo, D.L., 2018. Free radical scavenging, antimicrobial activities and effect of sub-acute exposure to Nigerian Xylopia Aethiopica seed extract on liver and kidney functional indices of albino rat. Iranian journal of toxicology, 12(3), pp.51-58.
- Yusuf, A.A., Lawal, B., Abubakar, A.N., Berinyuy, E.B., Omonije, Y.O., Umar, S.I., Shebe, M.N. and Alhaji, Y.M., 2018. In-vitro antioxidants, antimicrobial and toxicological evaluation of Nigerian Zingiber officinale. Clinical Phytoscience, 4(1), pp.1-8.
- Umar, S.I., Lawal, B., Mohammed, B.A., Obiekezie, C.I., Adewuyi, A.H., Babalola, S.B. and Ariyeloye, S.D., 2019. Antioxidant and antimicrobial activities of naturally occurring flavonoids from M. heterophylla and the safety evaluation in Wistar rats. Iranian Journal of Toxicology, 13(4), pp.39-44.
- Abimiku, R.H., Ngwai, Y.B., Nkene, I.H., Bassey, B.E., Tsaku, P.A., Ibrahim, T., Tama, S.C., Ishaleku, D. and Pennap, G.R.I. (2019). Molecular diversity and extended spectrum Betalactamase resistance of diarrheagenic Escherichia coli from patients attending selected health care facilities in Nasarawa state, Nigeria. International Journal of Pathogen Research. 3(1): 1 – 18.
- Makut, M.D., Agbonkhese, P.E. and Bello, A. (2018). Production of xanthan gum using Xanthomonas campestris isolated from some plants leaves in Keffi, Nigeria. Asian Journal of Biotechnology and Bioresource Technology. 3(4): 1 – 9.
- Makut, M. D., & Ibrahim, M. Z. (2021). Molecular and phylogenic identifications of potential herbicide degrading microorganisms from contaminated farmland in Keffi, Nasarawa State, Nigeria. AROC in Pharmaceutical and Biotechnology, 01 (01); 17, 25(1.0).
- Rana, B.M. and Raval, A.A. (2019). Isolation, production and characterization of the polysaccharide “xanthan gum” from Xanthomonas spp. Int. J. Curr. Microbiol. App. Sci. 8(5): 1019 – 1030.
- Tuli, H. S., Chaudhary, P., Beniwal, V., & Sharma, A. K. (2015). Microbial pigments as natural color sources: current trends and future perspectives. Journal of food science and technology, 52(8), 4669-4678.
- Yu, Y., Ma, J., Guo, Q., Ma, J., & Wang, H. (2019). A novel 3‐oxoacyl‐ACP reductase (FabG3) is involved in the xanthomonadin biosynthesis of Xanthomonas campestris pv. campestris. Molecular plant pathology, 20(12), 1696-1709.
- Palaniraj, A., & Jayaraman, V. (2011). Production, recovery and applications of xanthan gum by Xanthomonas campestris. Journal of Food Engineering, 106(1), 1-12.
- Murad, H. A., Mohamed, S. H., & Abu-El-Khair, A. G. (2017). Research Article Impact of Amino Acids, Nitrogen Source and Buffering System on Xanthan Yield Produced on Hydrolyzed Whey Lactose.
- Garcia-Ochoa, F. S., Casas, V. E., & Gomez, J. A. E., 2000. Xanthan gum: production, recovery and properties. Research review paper, Biotechnol Advences, 18, 1-31.
- Demirci, A. S., Palabiyik, I., Apaydın, D., Mirik, M., & Gumus, T. (2019). Xanthan gum biosynthesis using Xanthomonas isolates from waste bread: Process optimization and fermentation kinetics. LWT, 101, 40-47.
- Kumara, S. M., Khan, B. A., Rohit, K. C., & Purushotham, B. (2012). Effect of carbon and nitrogen sources on the production of xanthan gum from Xanthomonas campestris isolated from soil. Archives of Applied Science Research, 4(6), 2507-2512.